Unit 6 Topics
- Topics 6.1 and 6.1: DNA Structure and Replication
- Topics 6.3 and 6.4: Transcription and Translation
- Topics 6.5 and 6.6: Gene Regulation and Expression
- Topic 6.7, Parts 1, 2, and 3: Viruses
- Topics 6.7, Parts 4 and 5: Mutation and Horizontal Gene Transfer
- Topic 6.8: Biotechnology and Genetic Engineering
Essential Links
- The College Board’s AP Bio Course and Exam Description.
- My Condensed Version of the Course and Exam Description: takes the objectives, Enduring understandings, and key ideas of the 230-page CED and renders it into 40 pages.
- My AP Exam Review Outline: Takes the Course and Exam description and renders it into student (and teacher) friendly language.
- 2023-24 AP Bio Scope and Sequence Calendar: A spreadsheet that lays out the entire course.
Topics 6.1 and 6.2: DNA Structure and Replication
DNA Structure and Replication Learning Objectives
According to the College Board’s Course and Exam Description (see above for links) here’s what you should be covering about DNA structure and replication.
- Topic 6.1: DNA structure
- DNA is the molecule of heredity in most living systems. RNA can play that role in viruses
- Prokaryotes have circular chromosomes. Eukaryotes have linear chromosomes. Prokaryotes also have plasmids
- Base pairing rules: A w/ T; G w/ C. In RNA, A w/ U
- Topic 6.2: DNA Replication
- DNA replication occurs in a 5’ to 3’ direction (with new nucleotides added at the 3′ end.
- The process is semi-conservative
- The process involves a team of enzymes: Helicase, topoisomerase, DNA polymerase, and ligase are involved
- Replication happens differently between the leading strand and the lagging strand.
Here’s how to teach this material.
Start with some discovery-learning about the structure of DNA
Your students have already learned a bit about the structure of DNA in Unit 1.
To kick off Unit 6, I like to do an activity about how the structure of DNA was discovered. To do that, I use an activity: Discovering the Structure of DNA. This activity presents the “clues” that were uncovered in the 1800s and 1900s that eventually enabled Watson and Crick to develop their double helix model of DNA. After each clue, students draw inferences. At the end of the activity, students create their own model of DNA with puzzle pieces that represent deoxyribose, phosphate groups, and the four nitrogenous basis. Sounds easy, right? In my experience, students (at least my former students) find creation of a correct model (like the one below drawn by a former student) quite challenging.
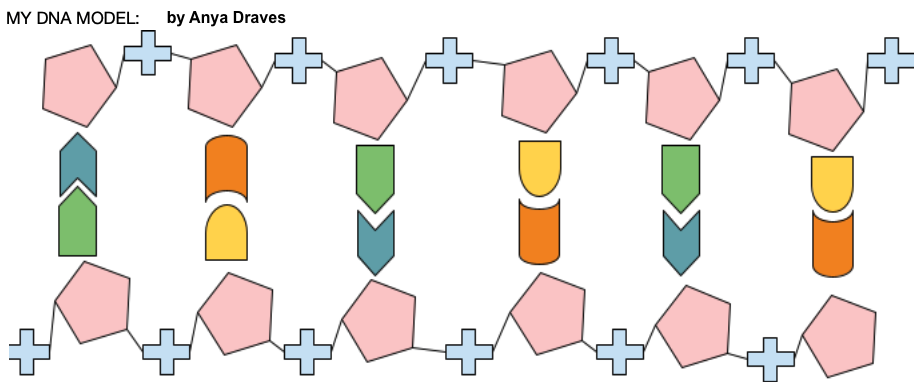
The activity takes about an hour, and it’s designed for students to do it in groups. If you do the paper version (which you can get by downloading the pdf at the link above), make the model pieces on cardstock, and save each group’s cutouts (so you can save time next year).
I liked to award “Nobel Prizes” to any group that gets their model right. My Nobel Prize was a gold star. If your students are like mine, you can get an AP Bio student to work pretty hard for a gold star!
Note that if you don’t want to worry about paper cutouts, then I have a completely electronic version of this activity (at the same link above).
DNA Structure and Replication Tutorials
Following that, you can send your students to learn-biology.com for these two tutorials:
- Topics 6.1 and 6.2, Part 1: DNA Structure (Review and Extension)
- Topics 6.1 and 6.2, Part 2: DNA Replication
which are accompanied by this student learning guide
The first tutorial gives your students mastery of diagrams like this:
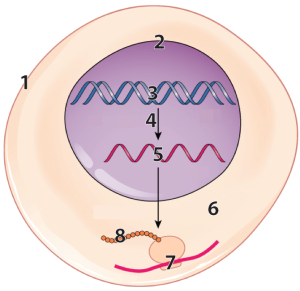
and related concepts: replication, transcription, and translation.
It also reviews material learned in Unit 1, and deepens students’ understanding of the structure of DNA, both at a big-picture level like this:
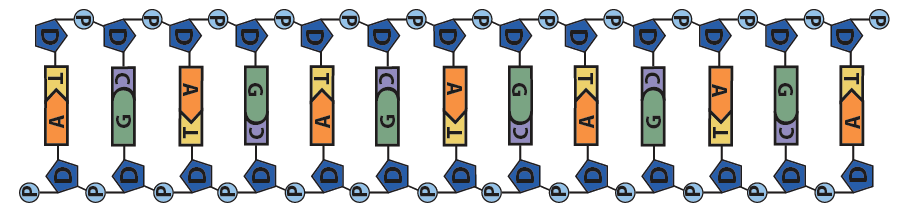
and also like this:
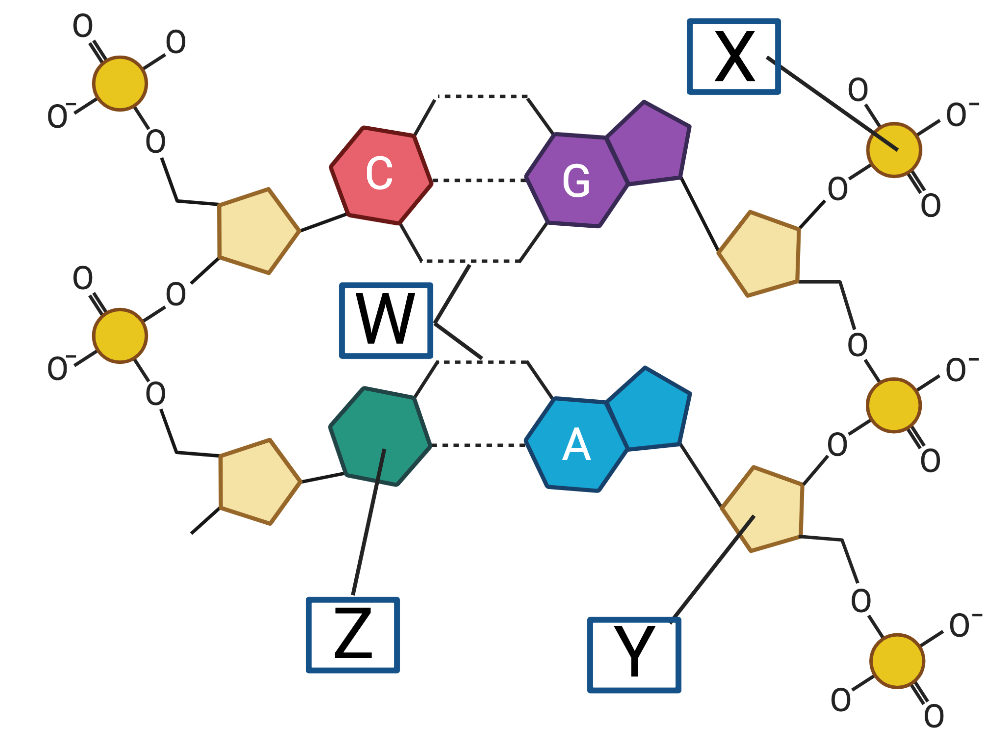
Concepts like base pairing rules, nucleotide structure, and hydrogen bonding, are all covered. Note that objectives related to plasmids and the differences between prokaryotic and eukaryotic DNA are covered later in Unit 6.
In my DNA replication tutorial, students learn about replication on a big-picture level, and then in a way that covers all of the required enzymes (except topoisomerase, so don’t forget to mention that to your students). By the end of the tutorial, your students will be able to talk their way through diagrams like this, explaining all the relevant enzymes.
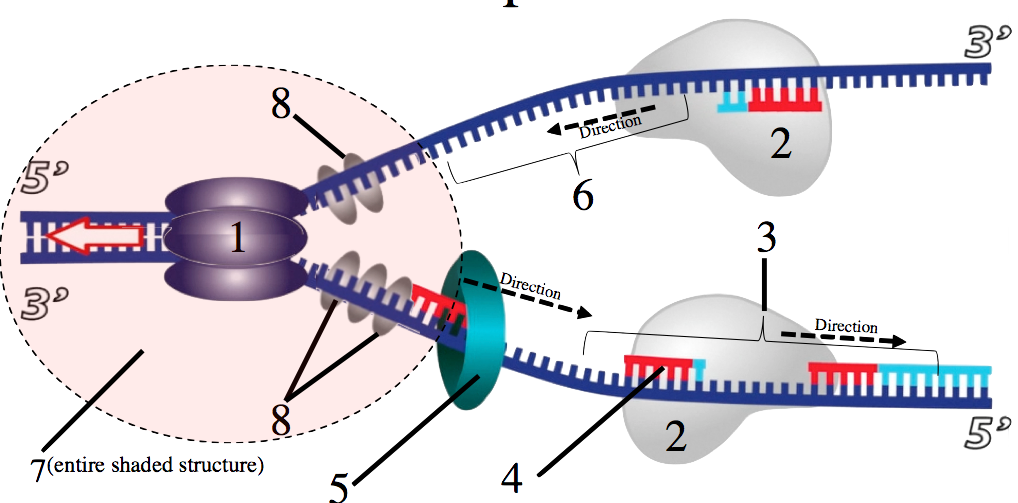
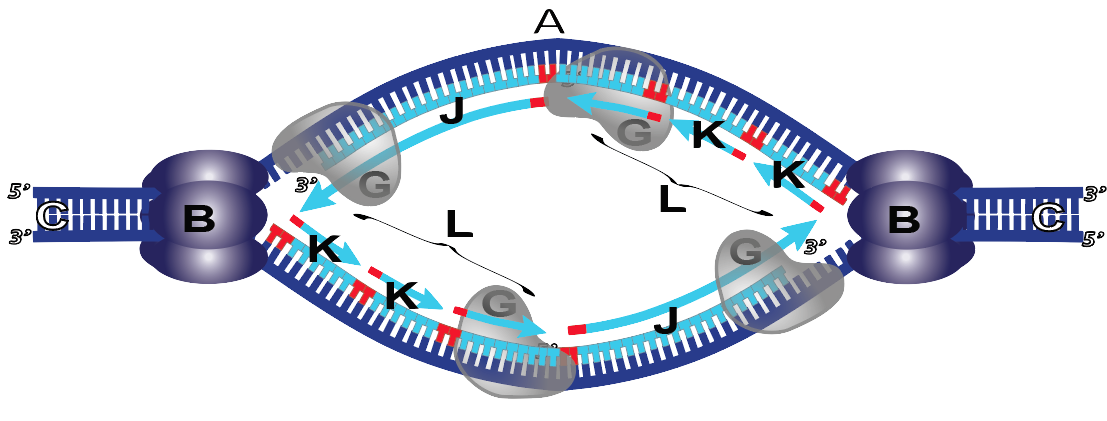
They’ll also have mastery of this one, contrasting the leading and lagging strands.
Other Resources
Here are a few other activities/resources for teaching DNA Structure and Replication
- Pulse-chase primer experiment (AKA Meselson-Stahl Experiment) HHMI. This is a fantastic way to walk your students through what has been called the “most beautiful experiment in biology.” Once you do this, your students will understand both what the semiconservative model of DNA replication is, and how it was verified.
- HHMI’s DNA Replication Computer Animation (see # 6 on my tutorial), or here on youtube. After you go over my tutorial with your students, you should show this much more detailed computer animation.
- Music Videos
- DNA, Fantastic! This is my music video that explains DNA structure. It was my first ever rap song, and my first collaboration with my music producer, Max Cowan. Note that this video, along with a Karaoke version, is also embedded into the DNA structure tutorial.
- DNA Replication: This music video explains the entire replication process, hitting almost every enzyme. It’s all done to a salsa beat, and the animations are very useful.
- DNA Replication Diagrams. This has pretty much every diagram of my website in a form that you can use to review with your students.
Topics 6.3 and 6.4: Transcription and Translation
Transcription and Translation: the Big Picture
When you’re teaching processes like transcription and translation, it’s easy to get lost in the details. So what’s important? Let’s look at transcription and translation from the perspective of biology’s four big ideas.
EVOLUTION: Transcription and translation are how changes in genotype by mutation or recombination manifest themselves as new phenotypes, which then get acted upon by natural selection. Think, for example, about COVID-19. it’s hard not to think of how viruses in the coronavirus family were randomly mutating and recombining until a viral RNA sequence arose that coded for a spike protein that was particularly good at binding with the ACE protein on the surface of epithelial cells in the human respiratory system. That was what made the spillover from bats to humans possible. It’s painful to think of this as an example of a successful phenotype, but that’s exactly what it is.
To go broader, this connection between information in nucleic acids and action in proteins is what life itself is all about. Years ago I was lucky enough to attend a lecture by Richard Dawkins in San Francisco. Dawkins was asked what life on other planets would be like. His response: some kind of genetic system for storing and transmitting information from cell to cell and from generation to generation. On Earth, that’s DNA. There would also be some system for manifesting this genetic information as an action system that would replicate those genes. On Earth, that’s to some extent RNA (which can itself be catalytic) but mostly it’s the protein that RNA codes for. On other planets, both systems could be chemically very different from what we have on Earth, but both will have to be there in some form.
INFORMATION FLOW: The flow of information in living things is what transcription and translation are all about. In terms of information, we see the central dogma at work. Information in a DNA template strand flows to pre-mRNA in a eukaryotic nucleus, then flows into the cytoplasm where ribosomes translate that message into protein. DNA triplets become mRNA codons, which become sequences of amino acids. To take this a step beyond the central dogma, this primary structure (the polypeptide sequence) has information to specify a specific 3D shape (which programs like Deep Mind can now predict).
Information flowing inward to organisms, tissues, or cells can impact the outward flow of information. To make this connection to your students, you can talk about how hormones like estrogen, testosterone, and growth hormone bind with cytoplasmic receptors, which then diffuse into the nucleus, activating transcription. This is a topic that we’ll cover later in Unit 6 when we look at operons, eukaryotic gene regulation, and development.
ENERGY FLOW: The synthesis of RNA sequences in mRNA, tRNA, and rRNA from RNA nucleotides is a big uphill push against entropy, and it doesn’t happen spontaneously. This endergonic reaction proceeds because RNA nucleotides come into the nucleus as triphosphate nucleotides (like ATP). RNA polymerases use the energy in those phosphates to synthesize strands of RNA. Similarly, a lot of ATP is required to power the formation of peptide bonds during protein synthesis. To help students make some connections that they might not make on their own, it’s a good idea to remind your students that this energy can be traced back to cellular respiration, powered by glucose (and other fuels), ultimately powered by the sun.
SYSTEMS: Transcription of RNA and protein synthesis are both processes that are deeply embedded in the structure of cells. But the system can be simplified to run in vitro, which you can read about here on Wikipedia. These cell-free systems are not new: Marshall Nirenberg used such a system to decipher the genetic code.
Transcription and Translation Learning Objectives
According to the College Board’s Course and Exam Description (linked above), here’s what you should be covering in relationship to transcription and translation. Note that I’ve rewritten these objectives into a student and teacher-friendly form.
TOPIC 6.3: Transcription
- Describe the flow of genetic information within cells (AKA the central dogma)
- DNA makes RNA makes protein
- Describe what happens during transcription
- RNA polymerase uses the sequence of DNA nucleotides in the template strand (AKA the noncoding strand, minus strand, or antisense strand) to synthesize complementary RNA.
- RNA polymerase binds at a promoter, a DNA sequence just upstream of the transcription start site.
- The binding of RNA polymerase can be regulated by transcription factors (see topics 6.5 and 6.6 below)
- The template strand varies depending on the gene that’s being transcribed.
- RNA polymerase synthesizes in the 5’ to 3’ direction as it reads DNA in the 3’ to 5’ direction
- Describe the roles and key features of the 3 types of RNA in protein synthesis
- mRNA carries information from DNA to the ribosome. Information is encoded in codons: 3 RNA nucleotides that specify a particular amino acid.
- tRNAs have an amino acid binding site and an anticodon (a sequence complementary to a codon). The specific binding of anticodon to codon ensures that the amino acid sequence in a polypeptide matches the sequence specified in mRNA
- rRNA is the catalytic part of the ribosome, connecting amino acids in a polypeptide chain.
Note that eukaryotic post-transcriptional processing of pre-mRNA to make mRNA is covered under eukaryotic gene regulation (below).
TOPIC 6.4: Translation
- Compare translation in prokaryotes and eukaryotes
- In all cells, translation occurs at ribosomes. In eukaryotes, some ribosomes are embedded into the rough E.R. (“bound ribosomes”) while other ribosomes float freely in the cytoplasm (“free ribosomes”)
- In prokaryotes, translation and transcription can occur simultaneously. In eukaryotes, transcription is in the nucleus, and translation in is the cytoplasm.
- Define the genetic code, describe its key features, and use a genetic code chart to predict the amino acid sequence that will be generated by a given sequence of mRNA.
- The genetic code is a set of 3-letter sequences of nucleotides called codons that code for specific amino acids.
- The code is redundant, with many codons coding for the same amino acid.
- The code is nearly universal, with only a few variants throughout the living world.
- The code includes punctuation: codons that signal for translation to start and stop.
- Describe the process of translation.
- Initiation: the small subunit of a ribosome binds with the start codon. A tRNA with an anticodon matching the start codon binds, bringing the first amino acid. The large subunit binds the small subunit.
- Elongation: As specified by the codon on the mRNA, tRNAs with the designated amino acid bind with the mRNA at the ribosome. The ribosome catalyzes a peptide bond between the newly arrived amino acid and the growing polypeptide chain.
- Termination: when a stop codon is reached, a release factor causes the polypeptide to be released, and the ribosome dissociates from the mRNA.
Material about retroviruses and reverse transcription are covered in a series of tutorials about viruses (see below)
On Learn-Biology.com, we cover everything listed above in the tutorials below.
Interactive Tutorials
Learn-Biology.com covers transcription and translation through four tutorials.
- Topic 6.3. Transcription
- Topic 6.4, Part 1: The Genetic Code
- Topic 6.4, Part 2: Translation/Protein Synthesis
- Topic 6.4, Part 3 (extension): Protein Targeting to the Rough E.R.
These are accompanied by this student learning guide.
The first tutorial starts by teaching the differences between DNA and RNA, and the slightly different base pairing rules (A pairs with U, not T). Students learn the basics of transcription and will be able to explain diagrams like this one…
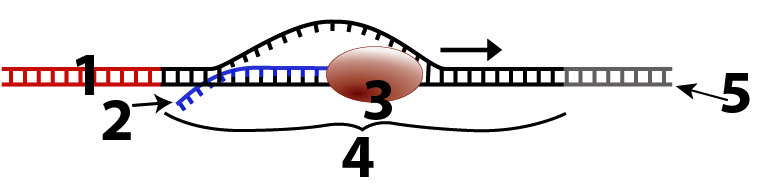
and this one:
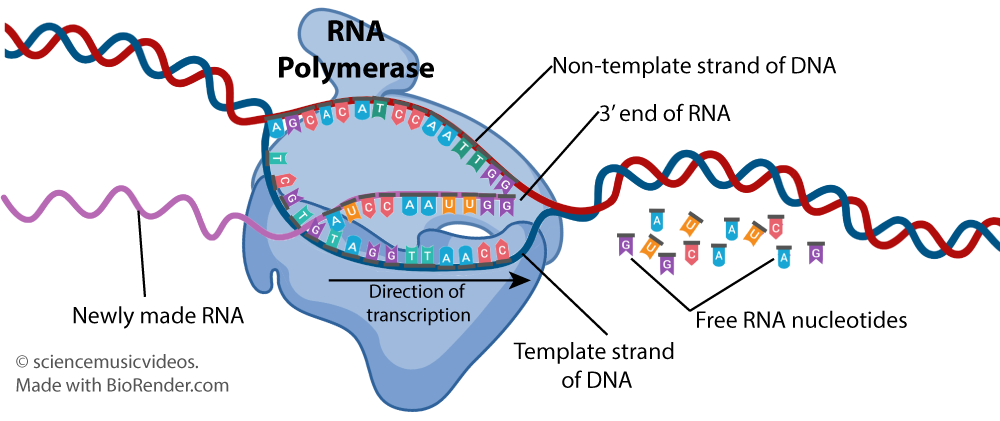
In the second tutorial, students learn how to use a genetic code dictionary:
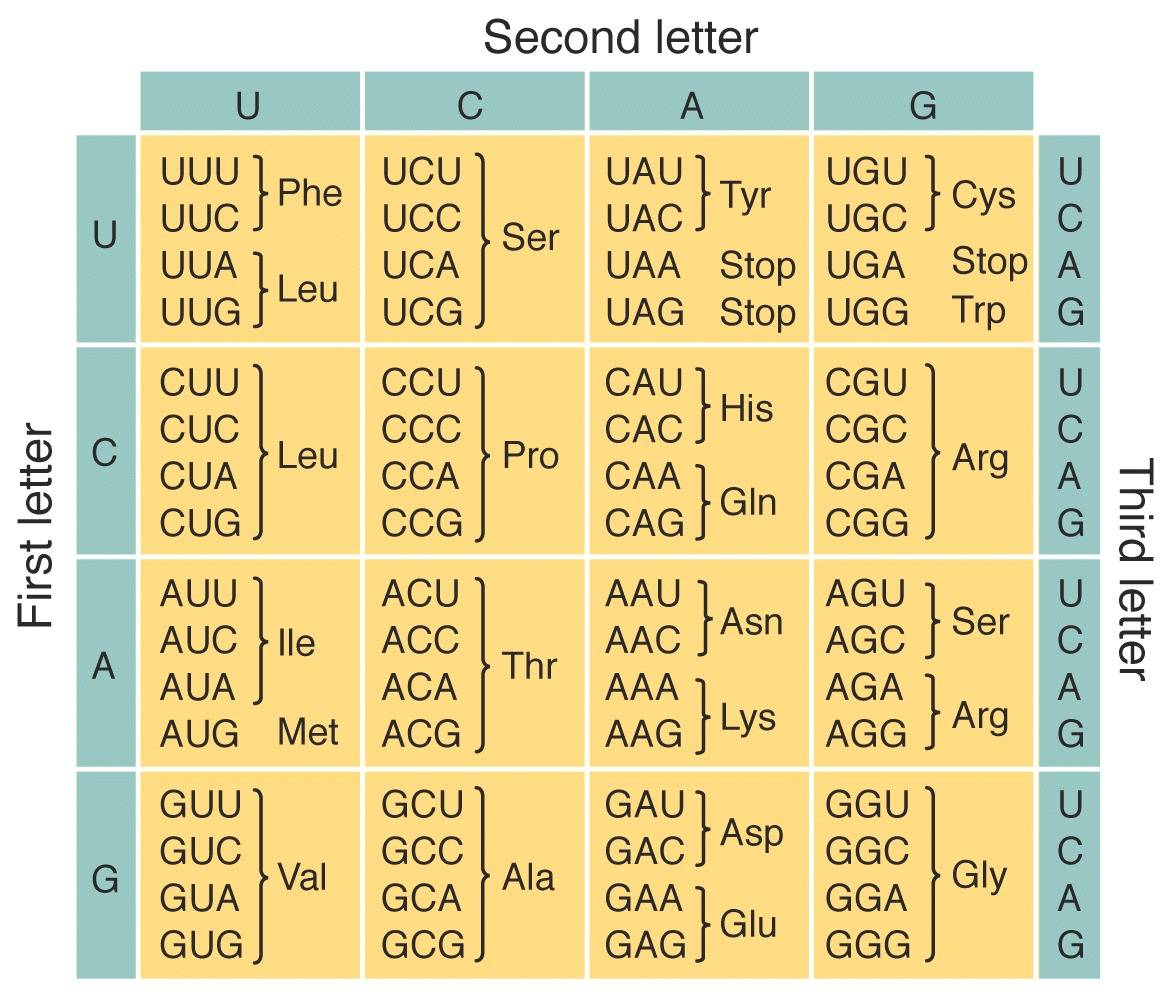
The third tutorial works through the entire process of protein synthesis, guiding your students through the various RNAs involved, and through diagrams like this:
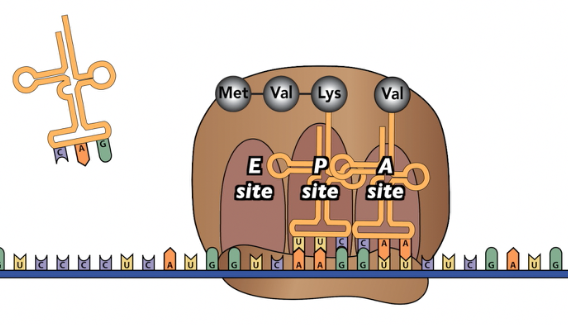
The last tutorial in this sequence goes outside of the standards to teach about protein targeting. The underlying question here is how ribosomes “know” whether to assemble their proteins freely in the cytoplasm, or as bound ribosomes attached to the Rough E.R. You can skip this if you’re pressed for time, but for me, this is a super-important feature of how cells work (and it only takes a few minutes).
Other Resources
Here are a few other activities/resources for teaching transcription and translation:
- Of all the music videos I’ve made, my Protein Synthesis Song is one of my personal favorites. It explains the whole process and has a great sing-along chorus. The video is embedded into the protein synthesis tutorial, and you can view it here on learn-biology.com.
- Flinn has two excellent POGILs on transcription and translation. They’re entitled “Gene Expression—Transcription”: and “Gene Expression—Translation.”
- Here’s a link to a guided lecture note-taking sheet. It has a lot of practice exercises about transcription, translation, and using the genetic code that you can use to check for understanding and for generating mastery.
- HHMI has a terrific series of activities that connect transcription and translation to the evolution of dark fur color in the rock pocket mouse. This is a follow-up to an activity I like to do in the first week of class (back in August) to teach my students how natural selection works. Now that your students understand transcription and translation, they can do this activity, which shows the molecular genetic basis of the MC1R mutation. Then you can use another HHMI activity about the Biochemistry and Cell Signaling Pathway of MC1R to connect the mutation to the expression of the alternative form of melanin (eumelanin) that occurs in dark-colored mice.
Topics 6.5 – 6.6: Gene Regulation and Expression
Topic 1: Operons
Here’s a student and teacher-friendly version of the College Board’s objectives related to operons. You can access the original wording under “Essential links” above.
- Define regulatory sequences.
- Regulatory sequences are DNA segments that control gene expression, usually by increasing or decreasing the transcription rate.
- Describe how prokaryotic cells use operons to control gene expression.
- Operons are clusters of genes under the control of a single promoter.
- The expression of operons is under the control of a regulatory protein, which binds to an operator region that is just downstream from the promoter.
- Describe how operons work as a regulatory system allowing bacteria to respond in an adaptive way to their environment. This should include a description of
- the Lac operon (an inducible operon)
- the Trp operon (a repressible operon).
While there’s nothing about operons that’s particularly difficult — it’s just one of many beautifully complex processes that we’re lucky enough to teach — there are a few important things to emphasize. One thing is what Sean Carroll in The Serengeti Rules calls double negative logic. In the lac operon, lactose shuts down a regulatory molecule, which otherwise would shut down the expression of the gene. The second thing is that the regulatory protein works through allosteric interaction with lactose. In fact, allosteric interactions, which you probably taught back in Unit 3 when you were teaching about enzymes, were discovered by Jacques Monod as he was figuring out how operons worked.
On Learn-Biology.com, we cover this material in one tutorial:
Topics 6.5 – 6.6, Part 1: Gene Regulation in Bacteria through Operons
By the end of the tutorial, your students will be able to explain what’s happening in this diagram of the lac operon.
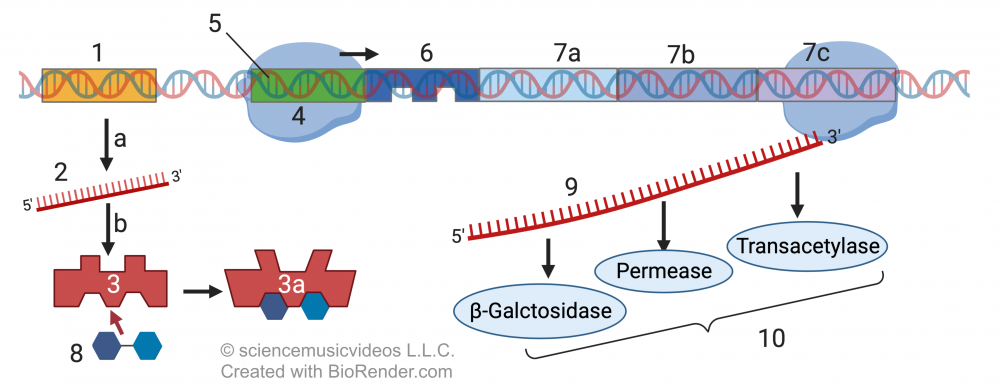
and this diagram of the trp operon
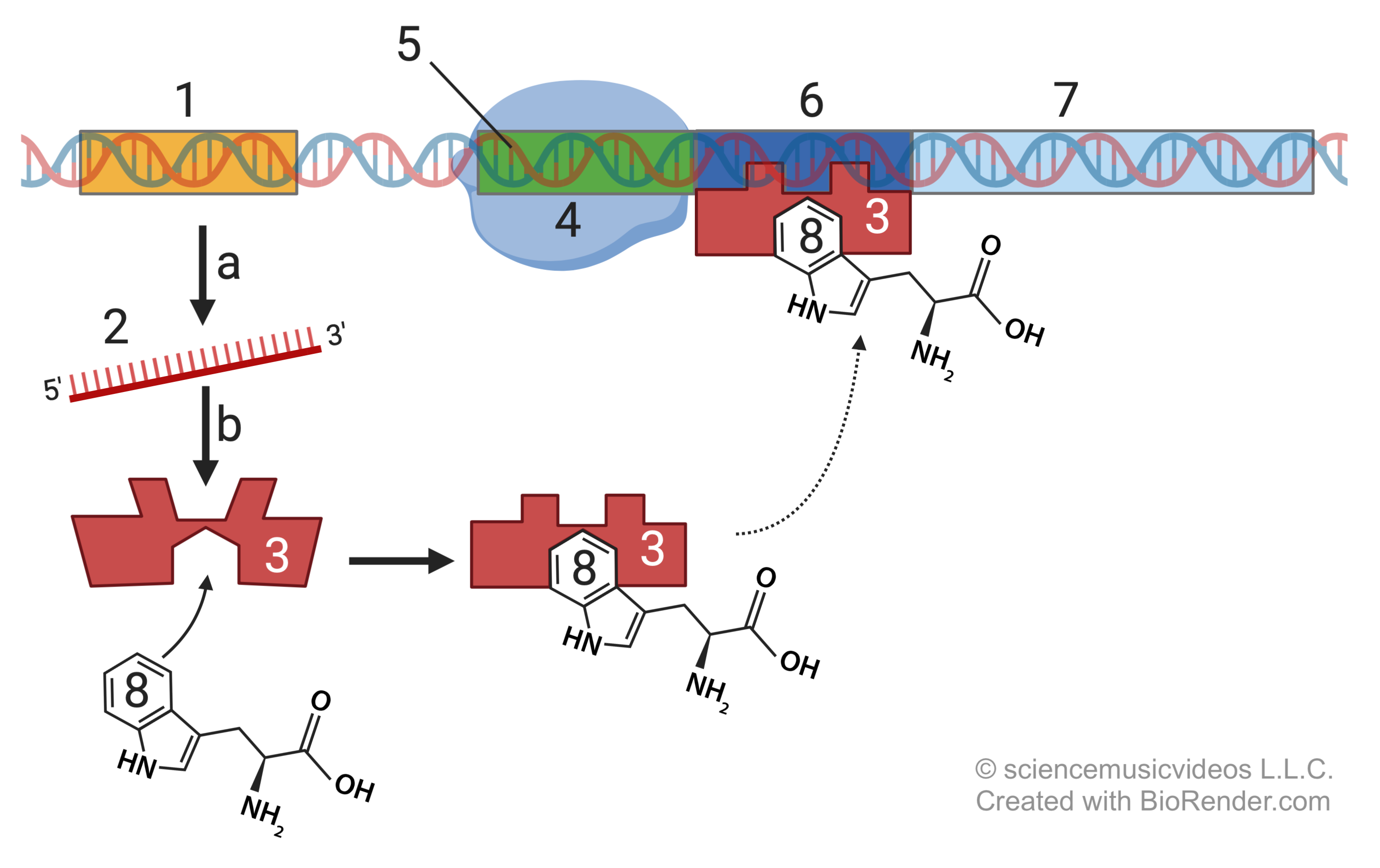
I recommend Flinn’s POGIL about operons. It’s entitled “Control of Gene Expression in Prokaryotes.” Here’s a copy of a worksheet that I use with my student to give them some additional practice, which can easily be adapted for direct instruction.
Topic 6.5, Part 2: Eukaryotic Gene Regulation
Once you’ve covered operons, you can move on to eukaryotic gene regulation. Here’s my rendering of the College Board’s objectives.
- Explain the role of transcription factors in regulating eukaryotic gene expression.
- Transcription factors are proteins that bind near or at the promoter to regulate the binding of RNA polymerase.
- Transcription factors can promote, block, or inhibit transcription.
- Negative regulatory molecules (including small RNAs) inhibit gene expression by binding to DNA and blocking transcription.
- Explain how the phenotype of a multicellular organism is determined by gene expression.
- All cells in a eukaryotic organism have the same DNA.
- Cells differentiate into specific tissues because they express genes for tissue-specific proteins.
- These tissue-specific genes are activated through the induction of transcription factors during embryonic development.
- Induction and gene activation unfold in a hierarchical sequence.
- Small RNAs also play a role in the regulation of transcription and translation.
- Explain how gene expression can be coordinated in eukaryotes
- In eukaryotes, genes in different tissues can share regulatory sequences that can be activated or repressed by transcription factors to coordinate gene expression.
- Define epigenetics.
- Epigenetics involves reversible modifications to DNA that influence gene expression without changing the DNA sequence.
On Learn-Biology.com, we cover this material in five tutorials
Topics 6.5 – 6.6, Part 2: Eukaryotic gene regulation through regulation of chromatin, Epigenetics
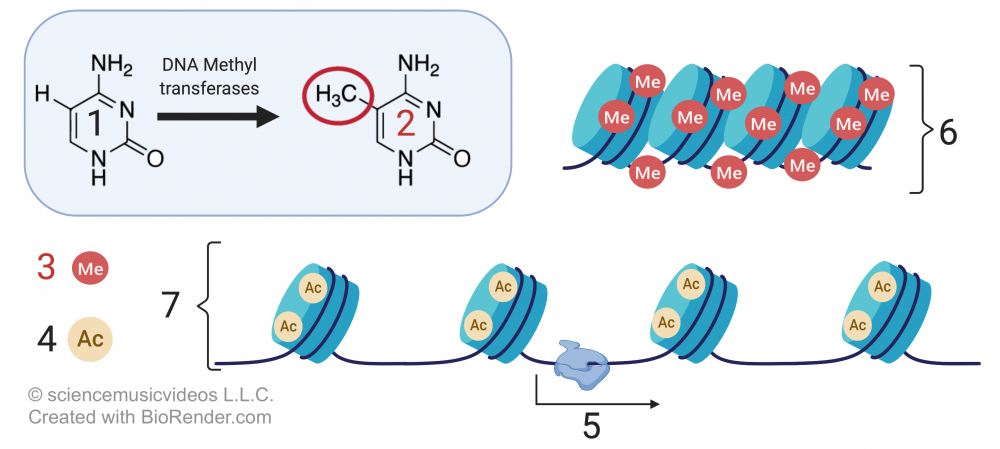
This module starts with a tutorial about the regulation of chromatin structure. The key thing I emphasize here is how most DNA in a cell is tightly wound up as heterochromatin, and not transcribed. A small proportion, by contrast, is euchromatin, which is available for transcription. These differences correspond to chemical modifications of either DNA or the histone proteins that package the DNA. A key diagram is this one. showing DNA methylation (6) and histone acetylation (7):
This can be pretty abstract unless you can fold it into a story…and fortunately, nature’s provided one. This is the story of X-chromosome inactivation, Barr Bodies, and how you can see that manifested in tortoiseshell cats.
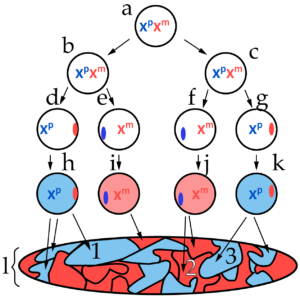
By the end of the tutorial, your students will be able to explain diagrams like this, which show the random pattern of X inactivation and Barr body formation that occurs early in female embryonic development…
…which, with a bit of imagination, gets you to this:

That kind of modification of DNA is a huge chunk of what epigenetics is: how modifications to DNA that don’t affect sequence control gene expression. The mind-blowing stuff is the intergenerational transfer of these modifications that can occur. I cover this through an excerpt about the Dutch Hunger Winter, and its intergenerational legacy.
Topics 6.5 – 6.6, Part 3: Eukaryotic gene regulation through control of transcription
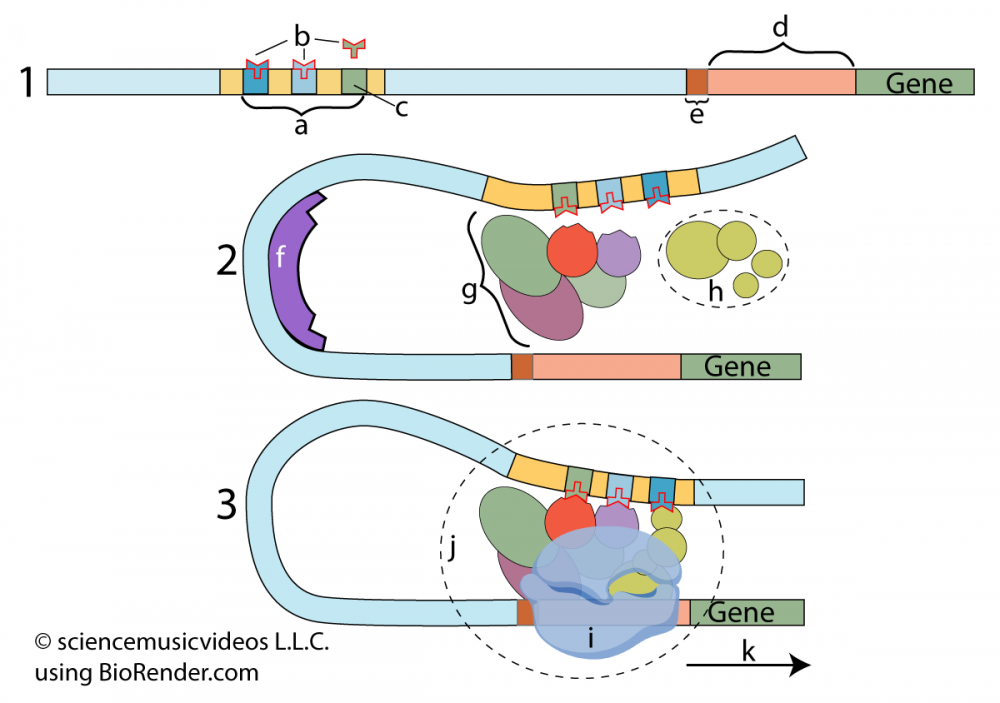
The second tutorial focuses on the regulation of transcription. In other words, for that portion of DNA that is transcribable, what determines when it gets transcribed? This focuses on the role of enhancers/regulatory switchers (at “a”) which interact with regulatory molecules (at “b”), leading to the formation of transcription initiation complexes (at “j”) that allow RNA polymerase to bind and transcribe DNA. These complexes look like this:
If the setup for transcription in Eukaryotes isn’t complex enough, there are post-transcriptional modifications required before eukaryotic RNA can become mRNA that gets translated into protein.
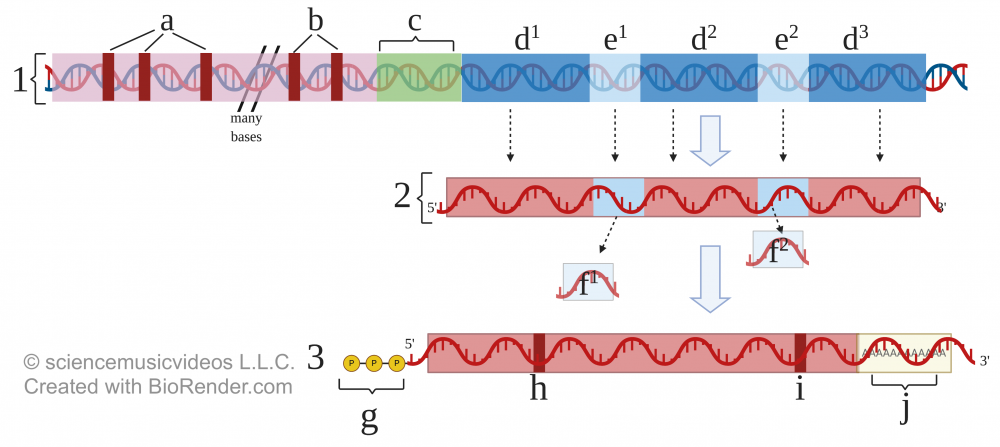
Through interacting with diagrams like the one above, students will learn about introns, exons, and other post-transcriptional RNA modifications.
We also focus on how the presence of introns within eukaryotic genes allows for exon shuffling, which increases phenotypic variety without enlarging the size of the genome.
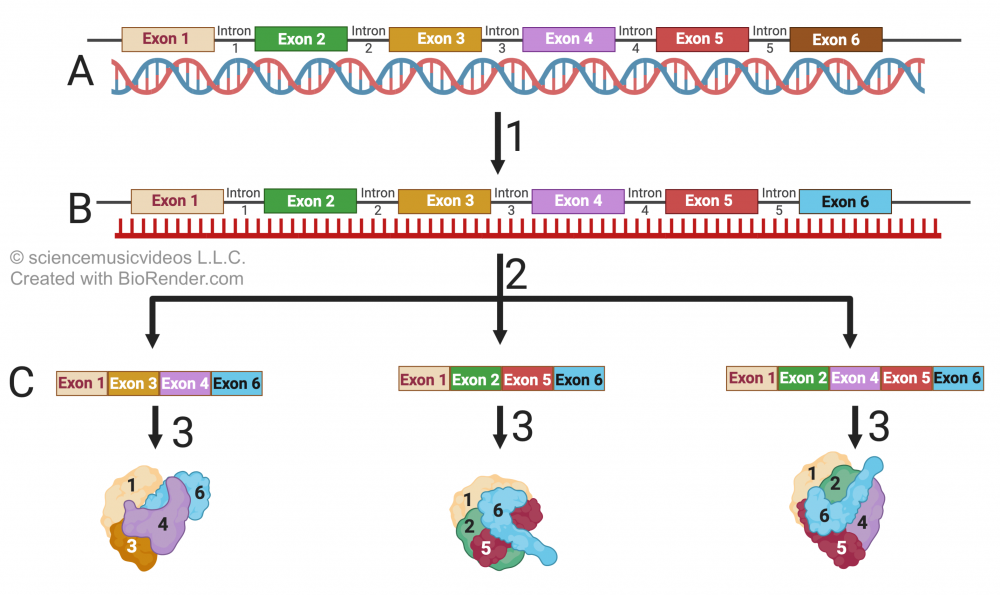
Topics 6.5 – 6.6, Part 4: Eukaryotic Gene Regulation — Coordinated Control Mechanisms, and microRNAs
This tutorial helps your students understand how the expression of genes in a variety of dispersed tissues throughout the multicellular body of a eukaryotic organism can be coordinated.
One mechanism is through common control elements, such as the drought response element in plants.
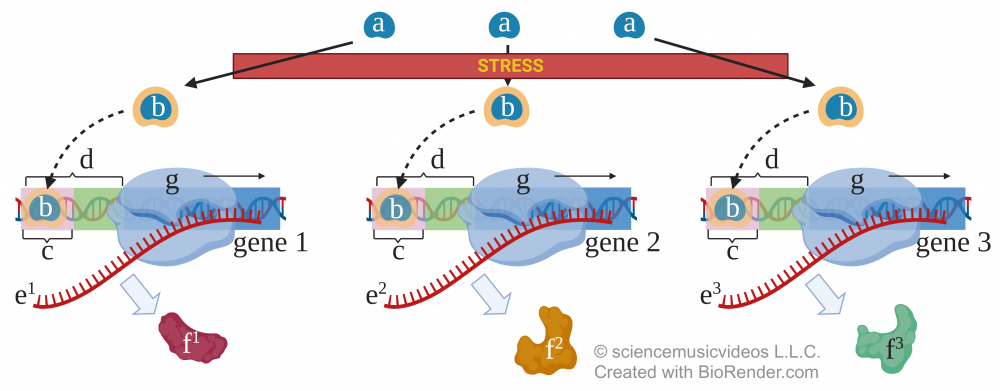
This sequence of DNA (shown in letter “d”, above) responds to the activation transcription factors (“a” to “b,” above) to turn on the expression of a variety of proteins (f1, f2, f3) that are part of the organism’s response to some environmental condition.
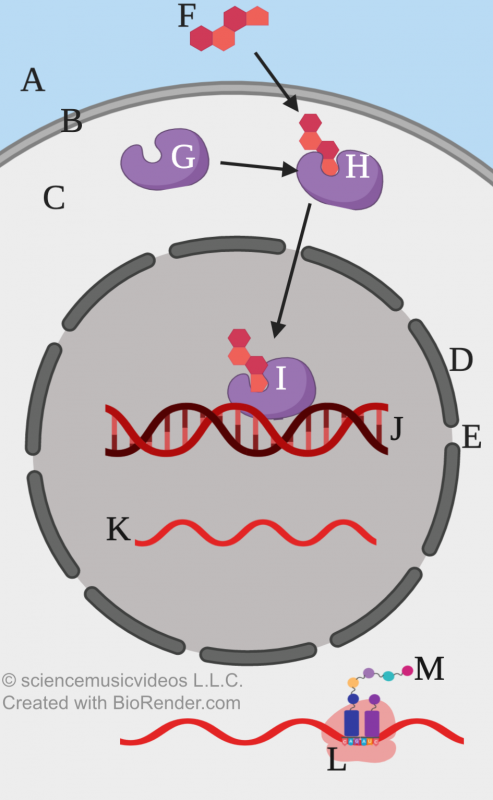 In animals, cytoplasmic receptors in a variety of tissues respond to steroid hormones (such as testosterone or estrogen) to bring about long-lasting changes in gene expression in a variety of tissues.
In animals, cytoplasmic receptors in a variety of tissues respond to steroid hormones (such as testosterone or estrogen) to bring about long-lasting changes in gene expression in a variety of tissues.
Topics 6.5 – 6.6, Part 5: Eukaryotic Gene Regulation and Development
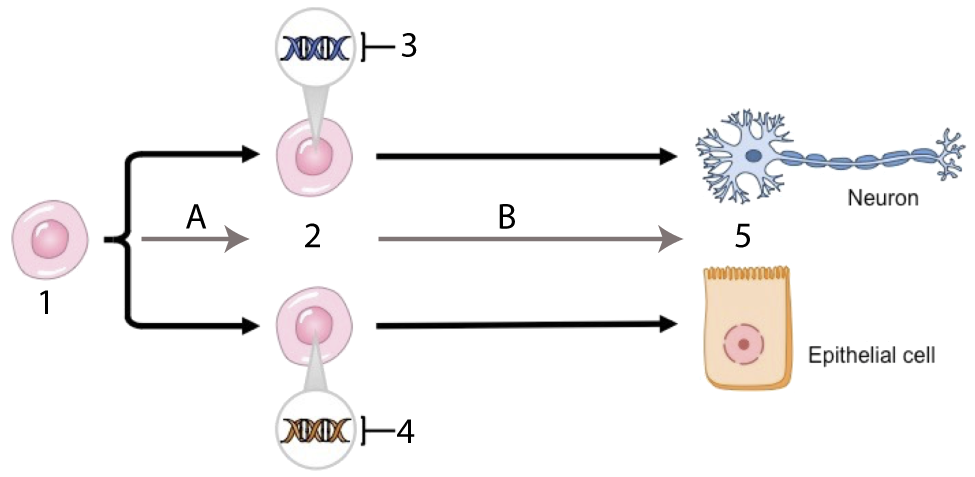 This tutorial hammers home the idea the differences in the tissues in a eukaryotic organism are not about differences in the genes in these tissues, but in which genes are expressed
This tutorial hammers home the idea the differences in the tissues in a eukaryotic organism are not about differences in the genes in these tissues, but in which genes are expressed
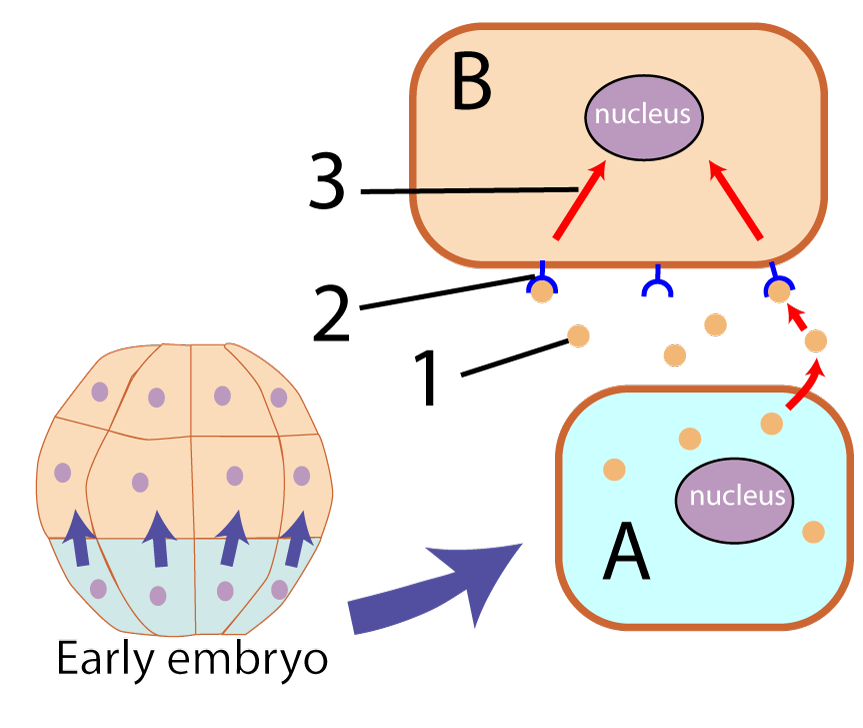 The tutorial continues with a discussion of induction (shown on the right), and an explanation of how gene expression in eukaryotes unfolds in a hierarchical sequence
The tutorial continues with a discussion of induction (shown on the right), and an explanation of how gene expression in eukaryotes unfolds in a hierarchical sequence
Topics 6.5 – 6.6, Part 6: Eukaryotic gene regulation and evolutionary change in the threespine stickleback
The last tutorial in Topics 6.5 to 6.6 looks at the intersection between gene regulation and evolutionary change, as seen in the Threespine Stickleback.
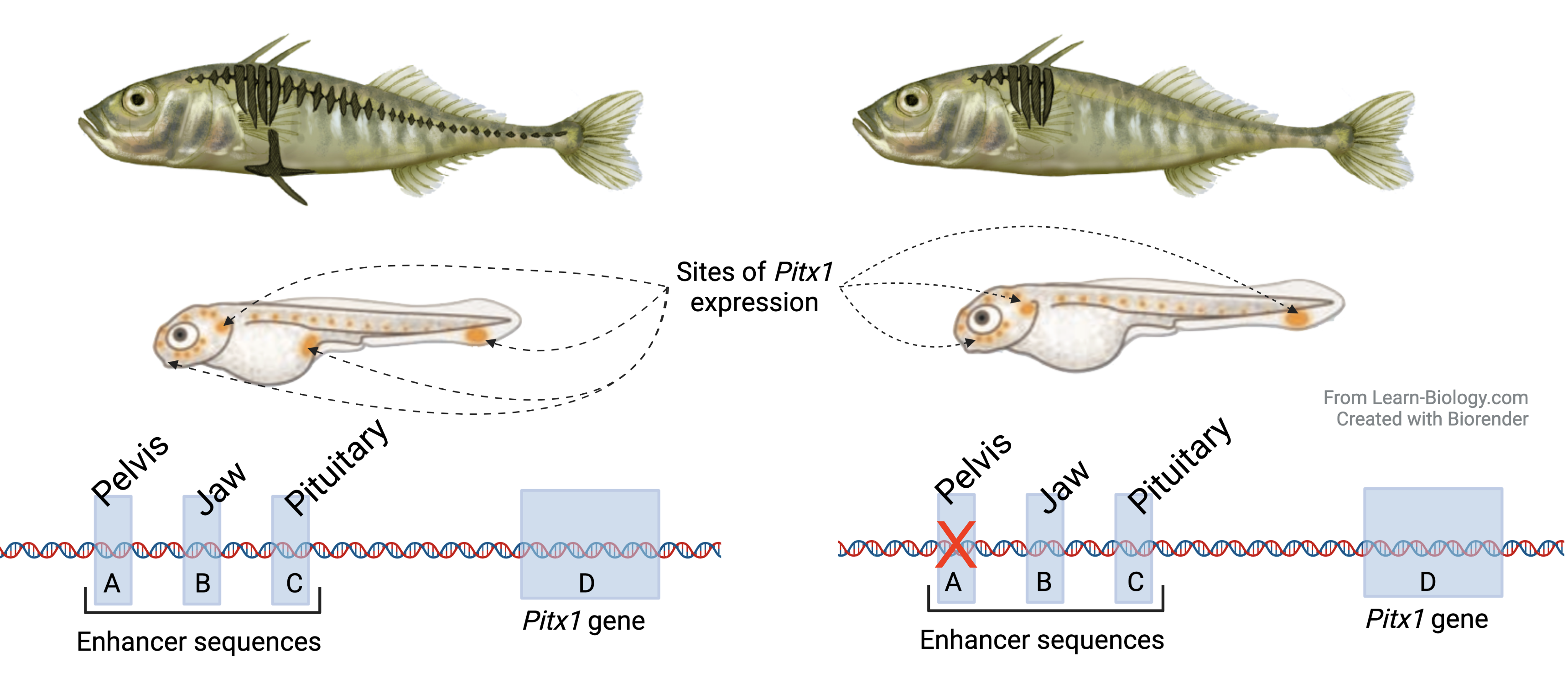
The difference between the spiny and spineless forms of these fish isn’t about the protein-coding Pitx1 gene, but mutations in the upstream pelvic enhancer. This tutorial (based on a video from HHMI’s Making of the Fittest series) is a fabulous preview of the material that you’ll be teaching about in AP Bio Unit 7, evolution.
Other Resources
HHMI has two activities related to gene regulation and evolutionary change in the three-spine stickleback. You can use them with the interactive module about this topic immediately above.
The first is HHMI’s stickleback virtual evolution lab. This is a multi-day simulated lab involving the examination of phenotype in both living and fossil sticklebacks, and then exploring the underlying genes that underlie the difference between the spiny and spineless forms.
The second involves showing the video Evolving Bodies, Evolving Switches, and then following it with this classroom activity: Modeling the Regulatory Switches of the Pitx1 Gene in Stickleback Fish. You can do this in one classroom period. Both the video and activity are highly recommended.
Finally, this video shows a very cool study from the University of Oregon about the pace of evolutionary change in three spine stickeback (spoiler: the transition from the spiny form to the spineless form can happen in just a few decades).
Topic 6.7, parts 1, 2, and 3: Viruses
Why teaching about viruses is so important
The word “virus” only shows up in two places in the Course and Exam Description. It shows up in topic 6.4 in the context of reverse transcriptase.
Genetic information in retroviruses is a special case and has an alternate flow of information: from RNA to DNA, made possible by reverse transcriptase, an enzyme that copies the viral RNA genome into DNA. This DNA integrates into the host genome and becomes transcribed and translated for the assembly of new viral progeny.
and it shows up in topic 6.7 (mutation) as
Related viruses can combine/recombine genetic information if they infect the same host cell.
But here we are, just on the other side of the COVID-19 Pandemic. The pandemic was a tragedy on so many levels…and it probably will be a milestone event in the lives of the students we teach for at least another decade. If our students aren’t going to learn about how viruses like SARS-CoV-2 work from us, they’re possibly never going to learn it. It’s our responsibility, as biology teachers, to provide accurate information to counter the lies and misinformation that many of our students have been exposed to.
In addition to that responsibility, teaching about viruses is also an opportunity. If your students are like the students I taught for the past 30 years, they’re interested in viruses. We’ve all been sickened by viruses! Viruses like the influenza virus, HIV and RSV are frequently in the news. And the way viruses work — the way that they hijack the cell’s molecular genetic machinery to reproduce themselves — makes for an amazing opportunity for you to deepen your students’ understanding of these processes.
Consequently, in the Calendar/Scope and Sequence I’ve shared with you, I’ve set aside three class periods for teaching about viruses. So please, teach about viruses.
Virus Learning Objectives
- Explain what a virus is, and how viruses straddle the boundary between living and non-living.
- Describe the basic structure of viruses, starting with phage, and including animal viruses.
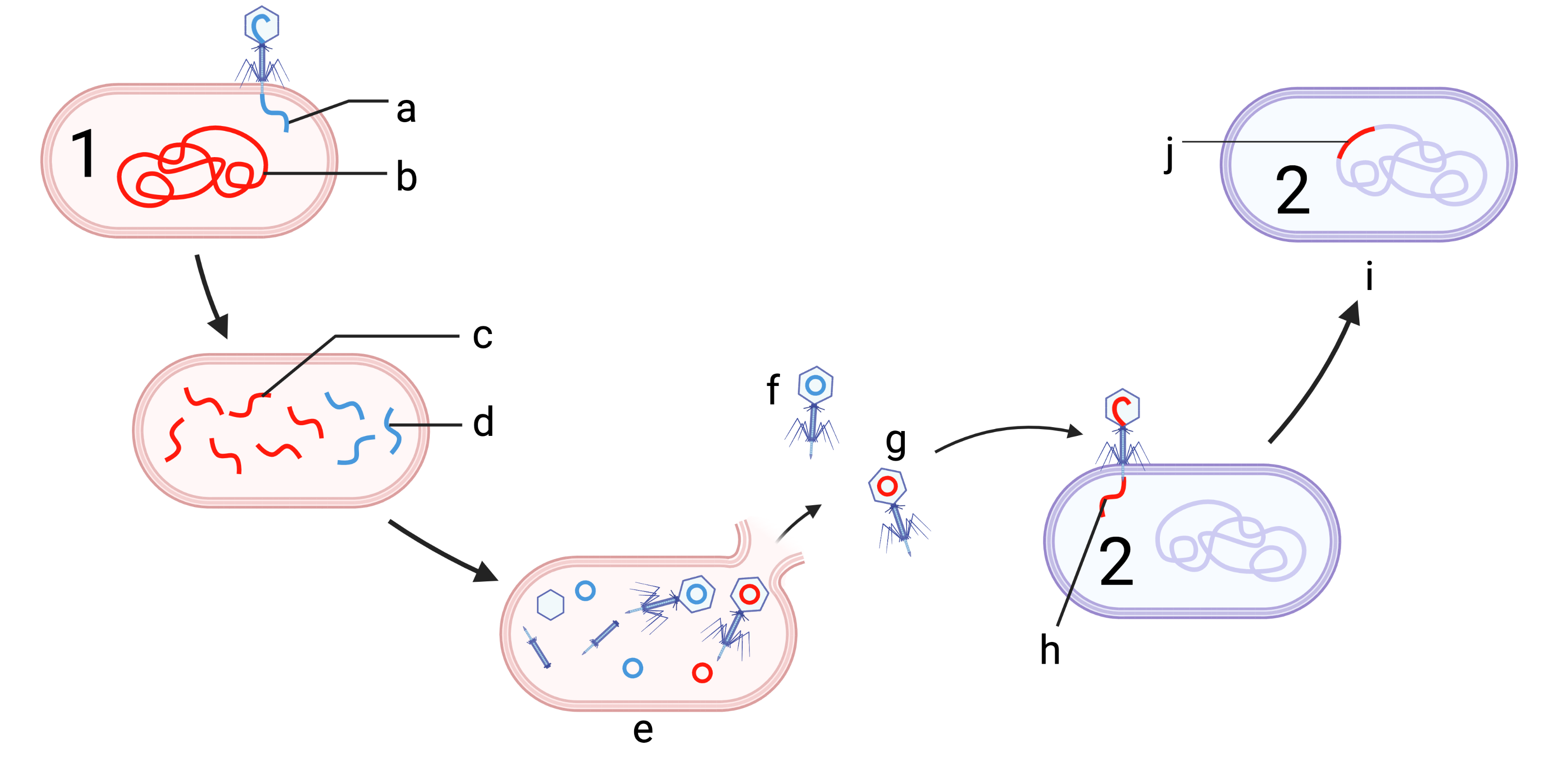
- Describe, compare, and contrast, the lytic and lysogenic cycles.
- As an illustrative example of a retrovirus, describe the life cycle of HIV. This covers the learning objective about retroviruses from Topic 6.4, above.
- Explain how viruses develop their own genetic diversity through recombination and mutation, and how they introduce variation into other species through transduction. This covers part of the learning objective from Topic 6.7 below.
- Describe the life cycle and overall biology of SARS-CoV-2.
- Explain how mRNA vaccines work to protect against viral infection.
- Explain how Rapid Antigen Tests work.
Viruses on Learn-Biology.com
Here’s an outline of what I teach in our Viruses tutorials. This material is embedded into Topic 6.7, and it’s supported by this student learning guide. I’m including relevant diagrams so you can get a quick sense of what your students will learn from our tutorials.
The first tutorial, Topic 6.7, Part 1: Virus Structure and Viral Life Cycles (Lytic, Lysogenic, and Retroviral), covers the following:
- What a virus is, and how viruses straddle the boundary between living and non-living.
- The basic structure of viruses, starting with phage.
- The lytic cycle
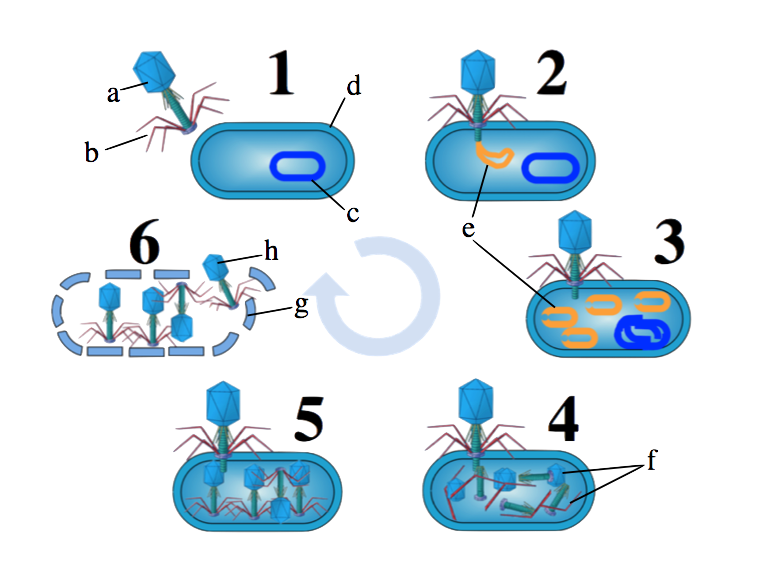
- The lysogenic cycle
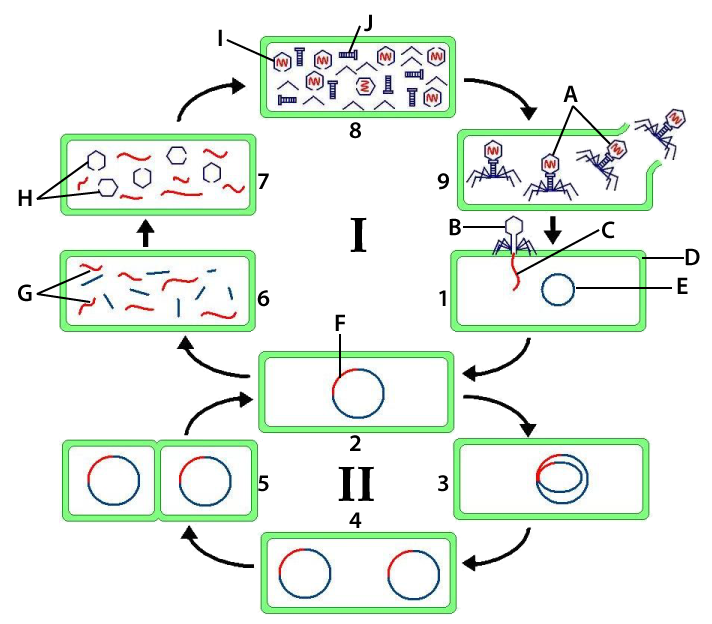
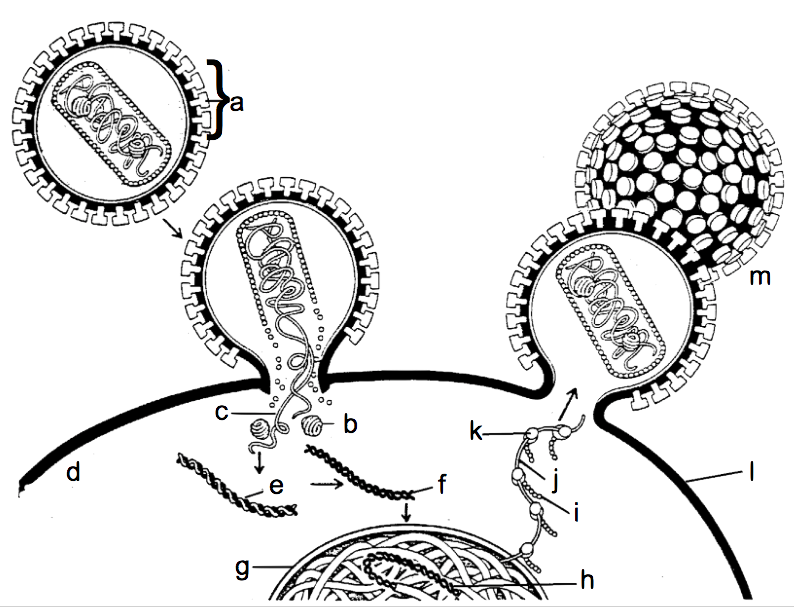
The life cycle of HIV (as an illustrative example of a retrovirus, which covers the retrovirus-related objective cited above)
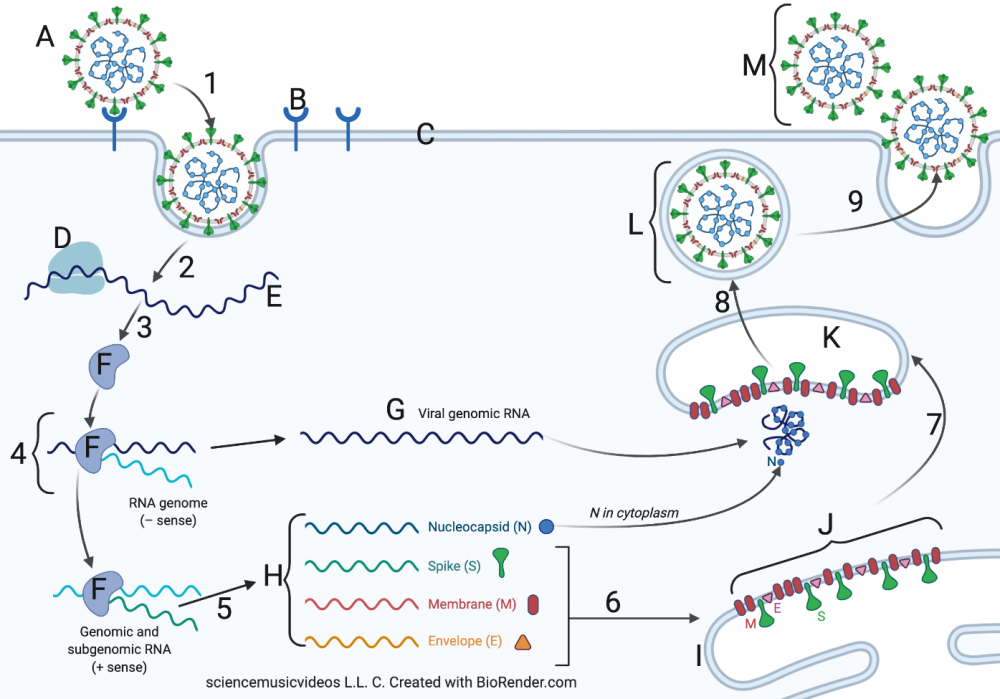
The second tutorial, Topic 6.7, Part 2: SARS-CoV-2 and Covid-19, covers the life cycle and overall biology of SARS-CoV-2
The third tutorial covers mRNA vaccines and Rapid Antigen tests.
Your students will gain mastery of diagrams like this:
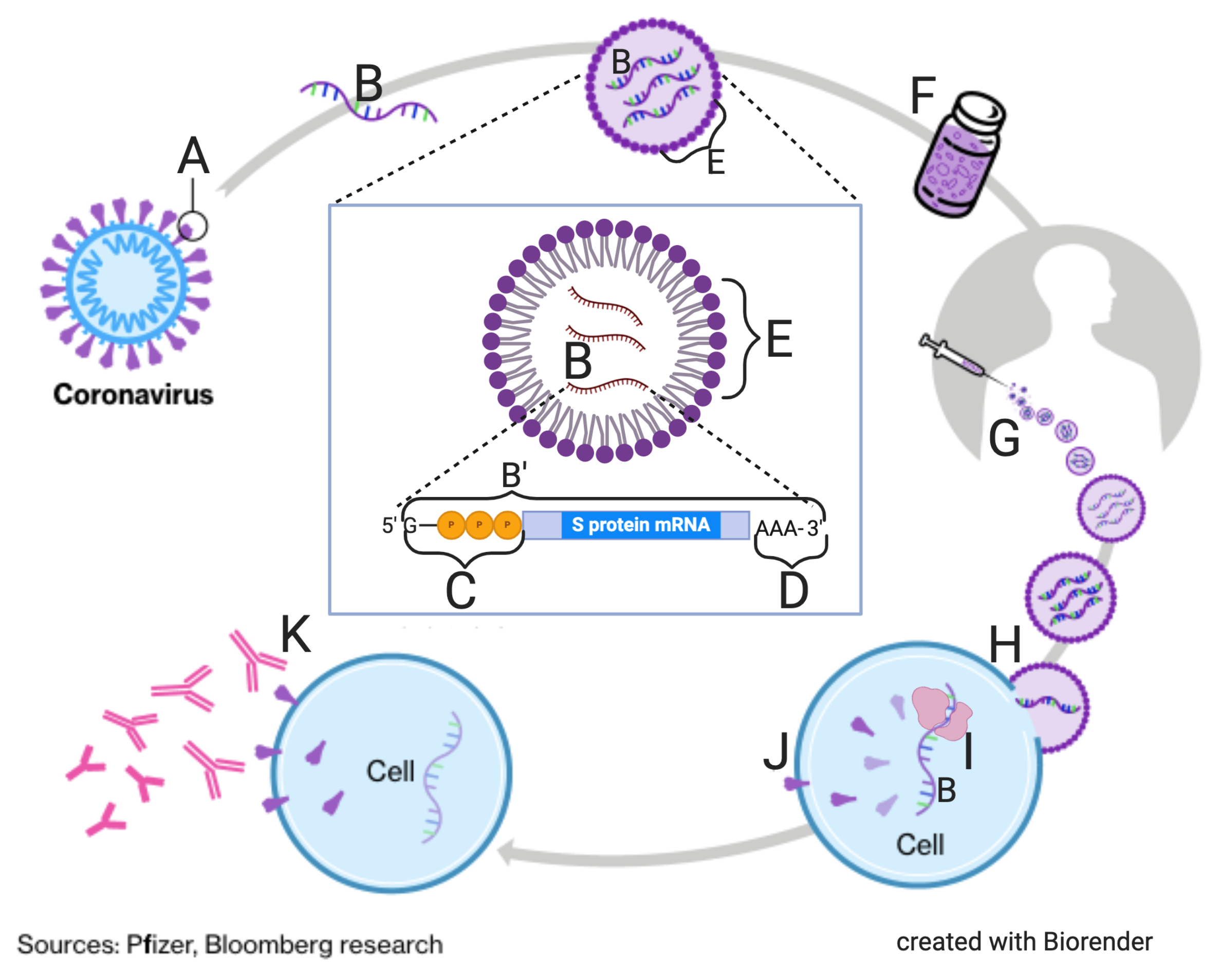
and this.
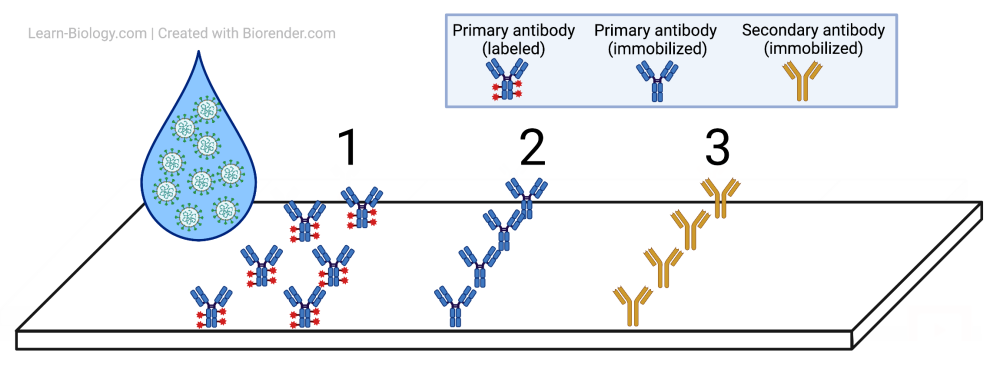
Again: I think it’s part of our mission as biology teachers to be promoting understanding of the novel coronavirus, and vaccines. I hope you’re on board with my battle against our common enemy Biology Confusion.
Additional resources for teaching about Viruses
- I have a list of resources at the bottom of the last tutorial. Giving your students a day to do open-ended research about COVID-19-related topics of interest and then sharing their results is a great use of class time.
- My music video, “I’m a Virus,” will help your students learn a lot of basic virus biology. It was written way before COVID, but it does include a stanza with visuals about the biology of HIV.
- Use this handout for direct instruction.
Topic 6.7, Parts 4 and 5: Mutation and Horizontal Gene Transfer
Mutation and Horizontal Gene Transfer Learning Objectives
If you want a look at the College Board’s Objectives for Mutations in my condensed outline, see the link above. What’s below renders these into a more student (and teacher) friendly form.
- Define mutation
- Compare and contrast somatic and germline mutations: where they occur, and what their consequences are.
- Describe various types of point mutations
- silent (no change in protein coded for by the mutated gene)
- missense (one amino acid is substituted for another one)
- nonsense (a stop codon is substituted for an amino acid)
- insertions and deletion errors leading to a frameshift (change in reading frame), causing extensive missense (or nonsense).
- Explain how mutations come about.
- Causes can include radiation, reactive chemicals, or errors in DNA replication or DNA repair.
- Explain how mutations can be harmful, beneficial, or neutral.
- Harmful: result in a protein that is nonfunctional or harmful (illustrative examples include mutations in the CFTR gene leading to cystic fibrosis)
- Beneficial: improves the function of a protein (illustrative examples include the MC1R mutation that leads to adaptive melanism, or the mutation leading to lactase persistence in certain human populations).
- Neutral: the resulting protein is the same (silent mutation) or similar (because of the type of amino acid substitution).
- Explain the overall importance of mutation to evolution.
- Mutation provides the raw material upon which natural selection acts.
- Connect the events of mitosis or meiosis to mutation (see related topics in Unit 5)
- Changes in chromosome number (polyploidy) resulting from mitosis or meiosis can create new species.
- Changes in chromosome number caused by nondisjunction during meiosis result in a variety of human developmental disorders (Down syndrome), or chromosomal differences (Turner Syndrome)
- Define horizontal gene transfer, and describe various types of mechanisms of horizontal gene transfer
- Horizontal gene transfer is the uptake of genetic information (as opposed to the inheritance of genetic information from a parent.
- It occurs primarily in prokaryotes and viruses via transformation, conjugation, and transduction.
- When two viruses infect the same cell, their genetic information can be combined, leading to viral progeny with novel gene sequences
NOTE: Chromosomal mutations related to non-disjunction are covered in this tutorial in Unit 5. We cover phenomena like the evolution of antibiotic and pesticide resistance in this tutorial in our Evidence for Evolution Module.
Mutations and Horizontal Gene Transfer on Learn-Biology.com
We cover the objectives above in two tutorials on Learn-Biology.com
Our Mutation tutorial starts by explaining the point mutation that results in sickle cell anemia.
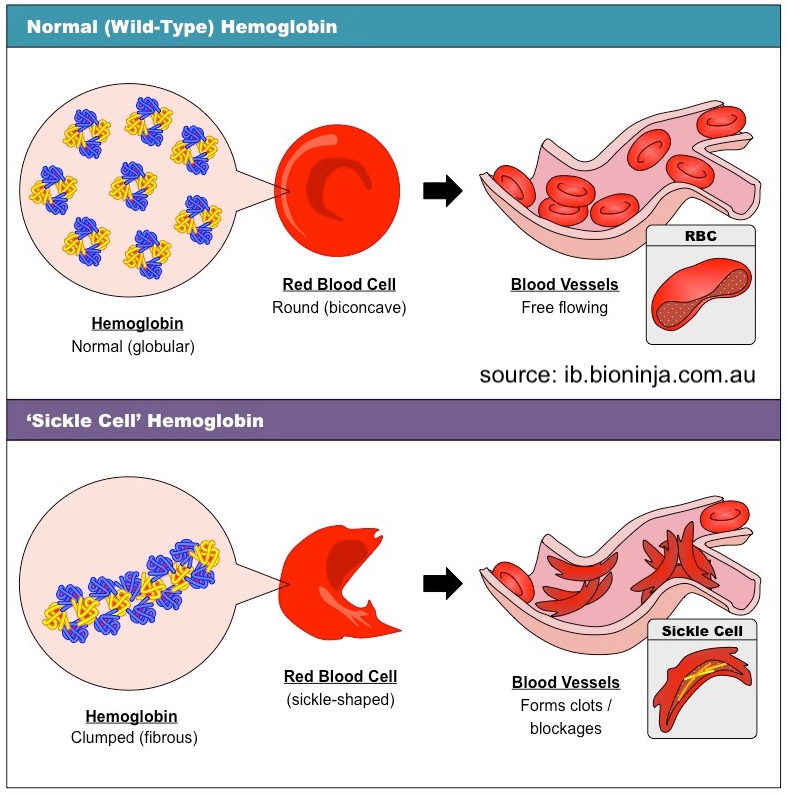
This is followed by material that teaches about
- the difference between germ-line and somatic mutations;
- the three types of point mutations (substitution, insertion, deletion);
- the results of mutation (silent, missense, or nonsense),
- some of the causes of mutation (mutagens/carcinogens), and
- how the consequences of mutation depend on the context.
Other Mutation-Related Learning Resources
Along with our tutorial, Flinn’s Genetic Mutations POGIL will help you solidify your student’s understanding of how mutations work. And if you haven’t already done these activities, you can work in any one of the HHMI’s activities on
- the molecular genetic basis of the MC1R mutation in the rock pocket mouse
- the Biochemistry and Cell Signaling Pathway of MC1R
- The mutation that causes the loss of pelvic spines in Three-spine sticklebacks. You can find these by going to https://www.biointeractive.org/classroom-resources and searching for “stickleback” and you’ll have many choices. But at the very least, I hope you have your students watch the video, and do this worksheet.
- the evolution of lactase persistence in several human cultures.
Horizontal Gene Transfer
Horizontal gene transfer is the movement of genetic material that occurs outside of reproduction.
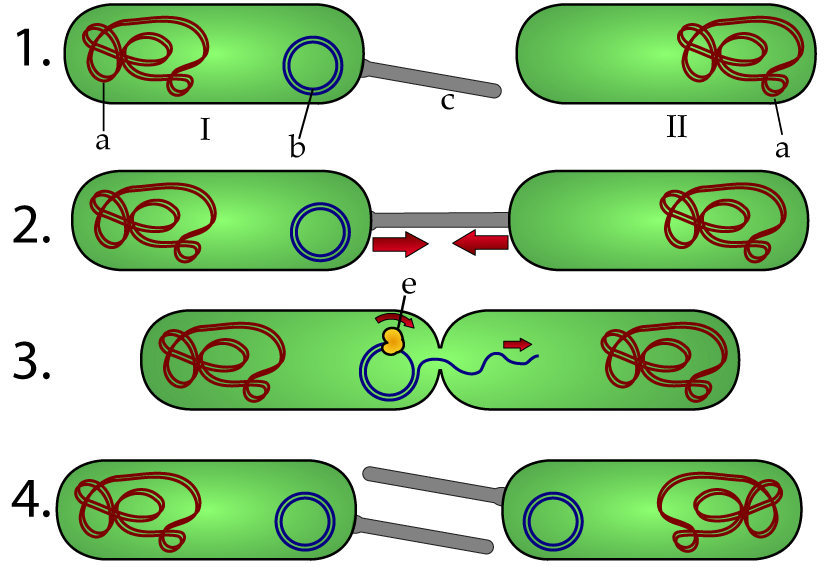
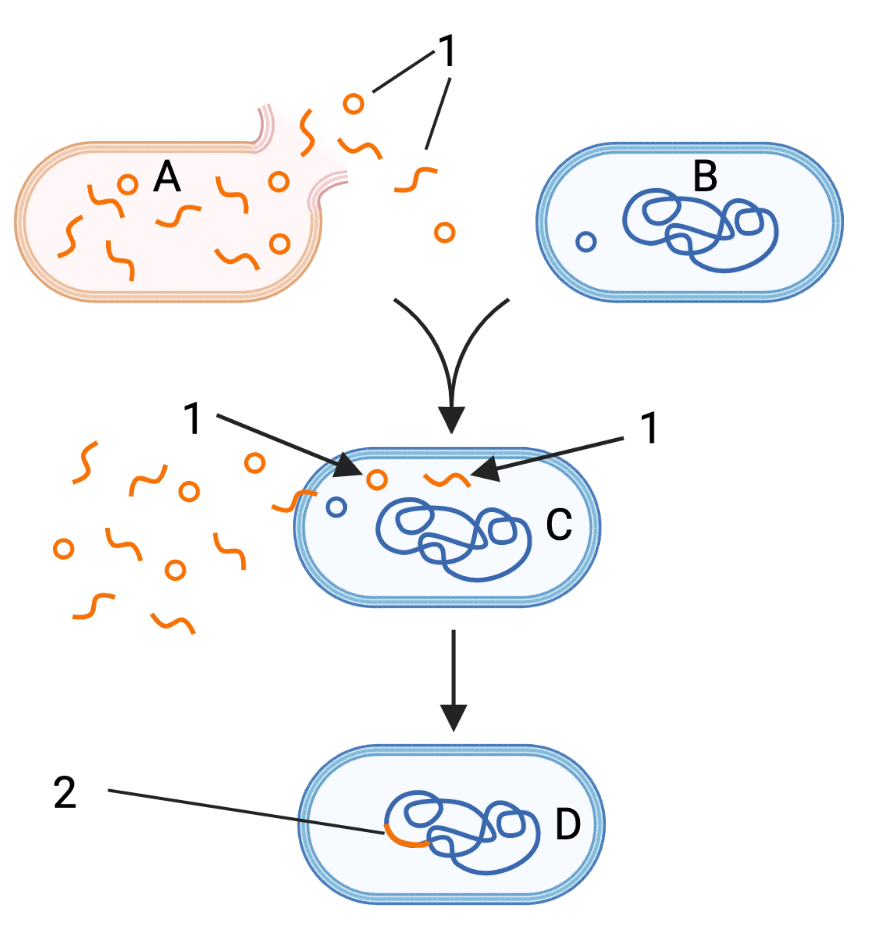
Our tutorial covers the three mechanisms of horizontal gene transfer shown below.
| bacterial conjugation | bacterial transformation | transduction |
 |
 |
 |
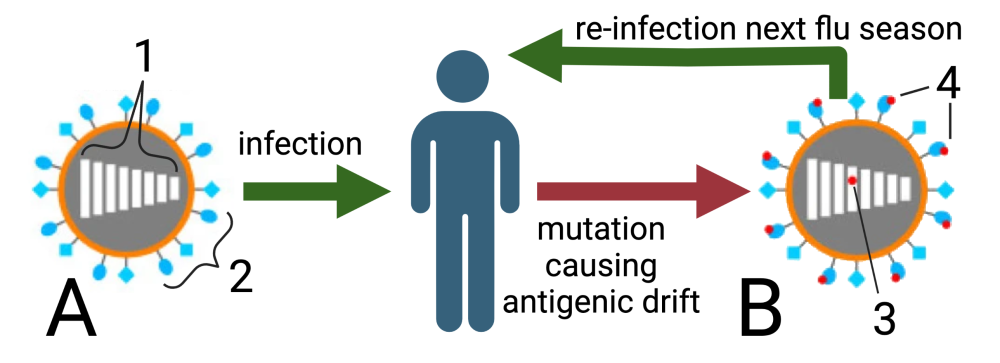
The tutorial ends with a comparison of antigenic drift and antigenic shift. The former process explains why we need to get a flu vaccine every year. The latter explains the occasional emergence of influenza strains that can cause worldwide pandemics.
| Antigenic Drift | Antigenic Shift |
 |
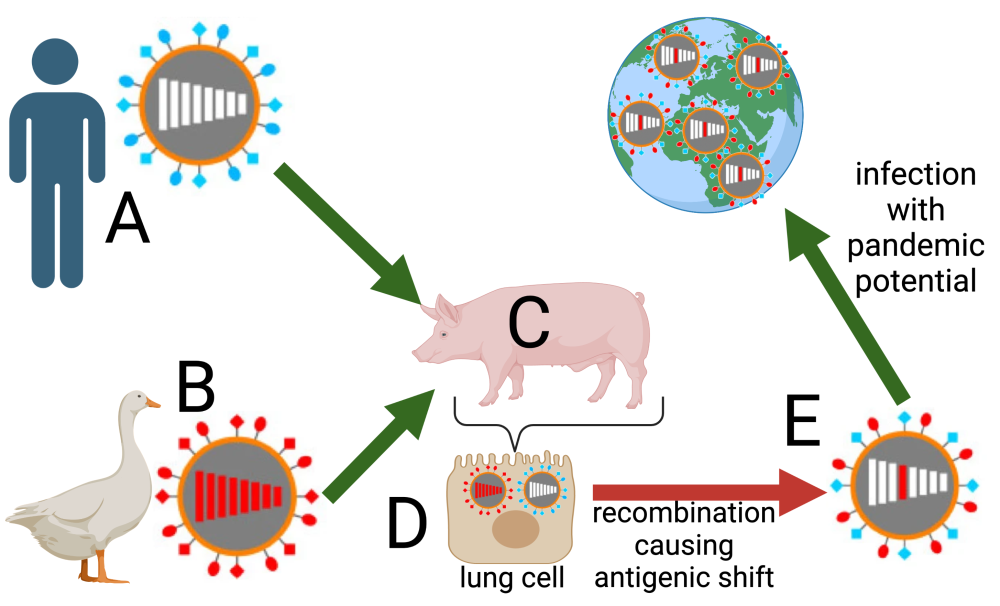 |
If you want to read a great summary of the differences between antigenic drift and shift, please take a look at this article from the NY Times (12/12/17): Why You Need the Flu Shot Every Year.
Topic 6.8: Biotechnology and Genetic Engineering
At Berkeley High School (my teaching alma mater), we spend about two weeks doing a series of biotechnology labs that are part of the AMGEN Biotechnology Experience. This is a series of labs that walks your students through:
- restriction digestion of plasmids
- gel electrophoresis
- bacterial transformation. This transformation is with a plasmid containing a gene for red fluorescent protein, and an arabinose-triggered operon. If you’ve done BioRad’s pGLO lab, this is very similar (and covers the same content).
Extensions allow you to do
- a ligation where you build the plasmid
- purification of gene products.
Amgen will train you for free, and then supply your school with all the needed supplies (consumables and hardware) for free. It’s amazing professional development for you, and a great lab experience for your students. I can’t recommend it highly enough.
In 2021, when the pandemic forced us into remote teaching, our team at BHS created this video. It shows the steps of the lab (carried out by my colleagues at Berkeley High School) and explains the science of the lab (explained by me). Please feel free to use it to orient your students to some key biotechnology techniques and concepts
Biotechnology and Genetic Engineering Learning Objectives (Topic 6.8)
You’ll find that what’s on Learn-Biology.com more than covers what the College Board, in their Course and Exam description, wants your students to know. Here’s an edited version of these objectives.
- Explain the basic goals of genetic engineering.
- Analyzing or manipulating DNA.
- Describe the basic method and purpose of electrophoresis
- Separating molecules of DNA, RNA, or protein according to size and charge, usually for analytical purposes.
- Describe the polymerase chain reaction (PCR)
- During PCR, DNA or RNA fragments are amplified (small amounts are made into larger samples that can be analyzed).
- Describe the purpose of bacterial transformation
- Introduces DNA into bacterial cells, usually to get these cells to express desired proteins (such as with genetically engineered insulin or clotting factor)
- Transformation is usually preceded by inserting novel genes into plasmids, which are then used as a vector to introduce these genes into bacterial cells where these genes can be replicated and expressed.
- Describe the purpose of nucleic acid sequencing
- Determining the order of nucleotides in DNA or RNA.
Biotechnology and Genetic Engineering Tutorials on Learn-Biology.com
Here are the tutorials I’ve written to help your students master the key ideas and techniques of biotechnology. Note that the first three tutorials lie squarely within the bounds of the AP Bio curriculum. The last tutorial ventures into explaining CRISPR…If you absolutely need to, you can skip it…However, I encourage you not to send your students into the world without understanding how this transformational technology works.
- Topic 6.8, Part 1: Genetic Engineering through Plasmids: Teaches your students how restriction enzymes, DNA ligase, etc. can be used to create genetically engineered products such as insulin.
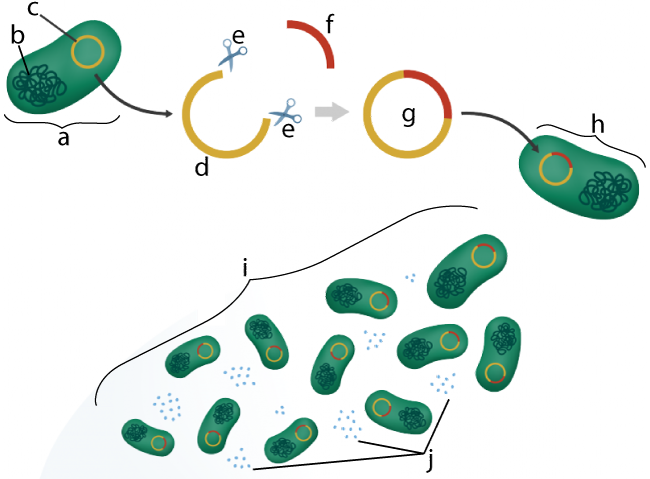
- Topic 6.8 Part 2: Gel Electrophoresis, Restriction Fragment Length Analysis, and DNA Fingerprinting: Teaches the basic principles of using restriction enzymes to create restriction fragments, which can then be sorted by size through electrophoresis.
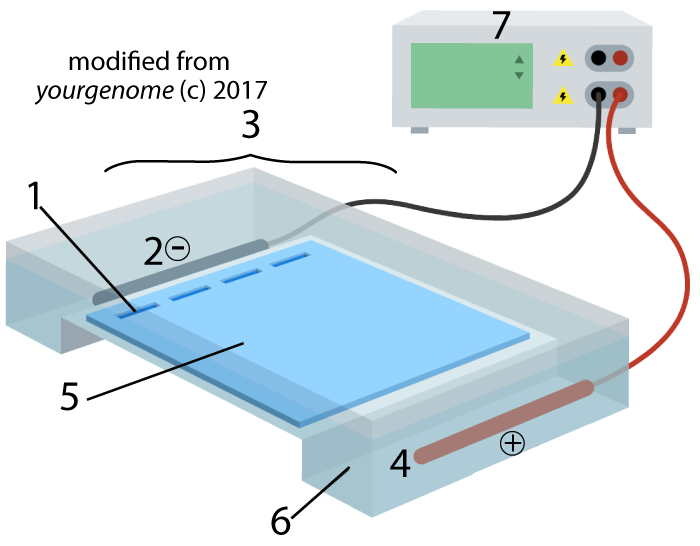
- Topic 6.8, Part 3: Amplifying DNA by PCR (the Polymerase Chain Reaction): A very short tutorial about the polymerase chain reaction.

- Topic 6.8, Part 4: A Few More Genetic Engineering Techniques to Know About: Sequencing and CRISPR: The sequencing section provides a 30,000-foot overview of DNA sequencing.
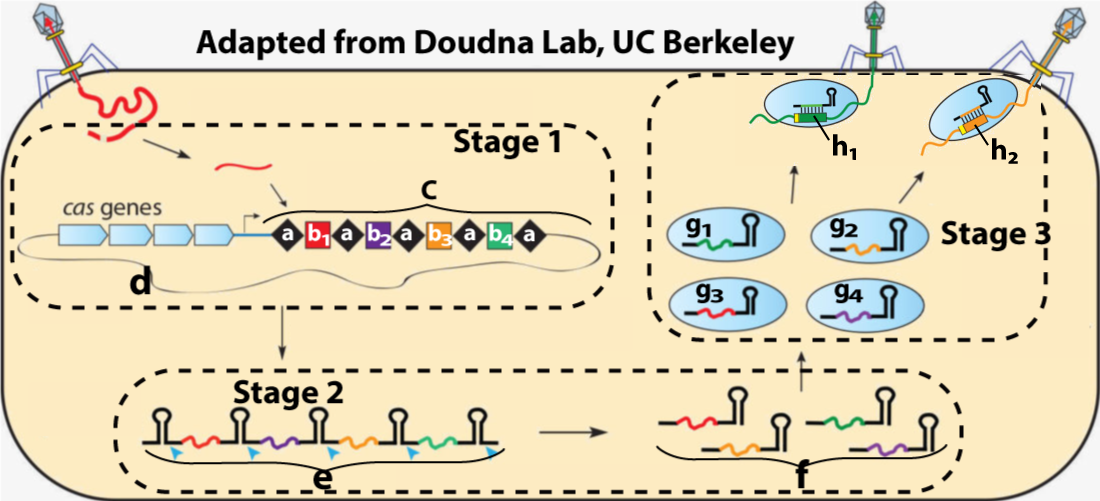
The section on CRISPR goes more into depth in helping your students understand this revolutionary gene editing system. Jennifer Doudna, who recently won the Nobel Prize for this work, lives in Berkeley (my hometown) and I was her son’s AP biology teacher. If you want your students to get to know her a bit better, you can show them this video of an interview I was able to do with her last year for a school fundraiser.
Additional Resources
If you can’t do any labs, then HHMI has several online simulations
- A bacterial identification lab that involves a simulation of PCR and electrophoresis.
- An interactive activity about CRISPR CAS-9
- A transgenic fly virtual lab
Summative Activities for Unit 6
To pull together Unit 6
- Use the AP Bio review materials on Learn-Biology.com. Your students can access these same materials on the Biomania AP Bio app, and results from both are recorded in your teacher dashboard. These review materials include
- Unit 6 AP Bio Review Flashcards
- Unit 6 AP Bio Review Multiple Choice Questions
- Unit 6 AP Bio Review Free Response Questions
- Click on Challenges
- The progress check questions for Unit 6 on AP Classroom.
