1. DNA Sequencing
In 1953, Watson, Crick, Wilkins, and Franklin established that DNA was the molecular language in which the code of life was written. In other words, the genetic difference between any two organisms lies in the sequence of A, T, C, and G nucleotides in their DNA. This initiated a worldwide effort — one which continues to this day — to figure out DNA sequences.
The process of taking a sample of DNA – anything from a small fragment to the entire genome of an organism – and figuring out the specific sequence of A, T, C, and G nucleotides that make it up, is called sequencing. For example, the table below shows the DNA sequence for a small part of a gene called NBN. NBN codes for a protein called nibrin, which has a variety of cellular functions, including repairing DNA damage. Take a moment to examine the sequence differences and similarities (something that’s mostly done by computers). Can you uncover any interesting patterns? Looking at sequences in this way is the basis of an entirely new field that fuses biology with information science that’s called bioinformatics, which has become a popular major in college. The application of bioinformatics to understanding the complete set of DNA within any organism has given rise to another new field: genomics.

DNA sequencing has become a mainstay of nearly every aspect of biology. On the most basic level, knowing the DNA sequence allows biologists to determine what proteins an organism can produce. In evolutionary biology, sequencing has become the main tool for inferring evolutionary relationships (a topic we’ll address in AP Bio Unit 7). Cancer biologists can sequence tumors to see what genetic mutations are causing cells to become cancerous. During the COVID-19 Pandemic, sequencing has been used to analyze the emergence of new SARS-CoV-2 variants. In forensics, sequencing is being used (along with DNA fingerprinting) to identify and exonerate suspects and resolve paternity disputes.
The first sequencing method was developed by Fredrick Sanger, and it’s explained in the video below.
Today, most sequencing is automated. Put your sample of DNA into a sequencer, and, depending on the size of the DNA sample, you’ll soon see a full readout of all of the As, Ts, Cs, and Gs that make up your sample of DNA. An entire human haploid human genome — over 3 billion bases — can be sequenced in less than an hour.
The details of Sanger’s method — which you’re entirely equipped to understand at this point in your AP Biology course — are fascinating, though beyond the scope of the AP Biology curriculum. If you’re interested in the details, you can learn about it in this college-level Learn-Biology.com tutorial.
2. CRISPR-Cas9: Introduction
If the first tutorial on this topic, we saw how recombinant DNA can be created and then inserted into the cells of an organism. But within the past decade, a much more flexible gene editing technique has emerged to change an organism’s DNA. It’s called CRISPR-Cas9. CRISPR is an acronym for clustered regularly interspersed short palindromic repeats. Cas9 stands for CRISPR-associated protein 9.
To get an overview of how CRISPR works, watch the video below and then answer the questions that follow.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-CRISPR-Cas9: Video Quiz (v2.0)”]
[h]Genome editing with CRISPR-Cas9: Video Quiz
[i]
[q] CRISPR first evolved as a system that bacteria used to protect themselves from attack from [hangman].
[c]IHZpcnVzZXM=[Qq]
[q multiple_choice=”true”] Cas9 is a nuclease. That means that it’s an enzyme that’s capable of cutting up
[c]IHBob3NwaG9saXBpZHM=[Qq]
[f]IE5vLiBUaGluayBhYm91dCB3aGF0IHZpcnVzZXMgaW5qZWN0IGludG8gY2VsbHMgd2hlbiB0aGV5IGF0dGFjayB0aGVtLg==[Qq]
[c]IHByb3RlaW5z[Qq]
[f]IE5vLiBUaGluayBhYm91dCB3aGF0IHZpcnVzZXMgaW5qZWN0IGludG8gY2VsbHMgd2hlbiB0aGV5IGF0dGFjayB0aGVtLg==[Qq]
[c]IA ==bnVjbGVpYyBhY2lkcw==[Qq]
[f]RXhjZWxsZW50ISBDYXM5IGlzIGEgbnVjbGVhc2UsIGFuIGVuenltZSBjYXBhYmxlIG9mIGN1dHRpbmcgdXAgRE5BLg==[Qq]
[q] CRISPR-Cas9 uses guide [hangman] to find matching DNA from a [hangman]. When it does, it makes a double-stranded cut in the viral [hangman], which disables the virus and keeps it from destroying the cell.
[c]IFJOQQ==[Qq]
[c]IHZpcnVz[Qq]
[c]RE5B[Qq]
[q]In recent years, scientists have engineered the CRISPR-Cas9 system so that instead of only cutting DNA from a [hangman], it can cut any targeted sequence of [hangman] in any organism.
[c]dmlydXM=[Qq]
[c]RE5B[Qq]
[q multiple_choice=”true”] After the Cas9 enzyme cuts up targeted DNA in a cell, the cell
[c]IGluaXRpYXRlcyBjZWxsIHN1aWNpZGUgcGF0aHdheXMgKGFwb3B0b3NpcykgdGhhdCByZXN1bHQgaW4gbHlzb3NvbWFsIHJ1cHR1cmUgYW5kIGV2ZW50dWFsIGNlbGwgZGVhdGgu[Qq]
[f]IE5vLiBBcG9wdG9zaXMgaXMgYSByZWFsIHBoZW5vbWVub24sIGJ1dCBjZWxscyB1c3VhbGx5IHNhdmUgdGhhdCBtb3ZlIGFzIGEgbGFzdCByZXNvcnQuIE5leHQgdGltZSwgY2hvb3NlIGFub3RoZXIgYW5zd2VyLg==[Qq]
[c]IHJlY29ubmVjdHMgdGhlIGJyb2tlbiBETkEsIGJ1dCBvZnRlbiBpbiBhIHdheSB0 aGF0IHJlc3VsdHMgaW4gbXV0YXRpb25zIHRoYXQgZGlzYWJsZSB0aGUgZ2VuZS4=[Qq]
[f]IFRoYXQmIzgyMTc7cyBleGFjdGx5IHJpZ2h0LiBDZWxscyByZXNwb25kIHRvIEROQSBicmVha3MgdGhyb3VnaCBETkEgcmVwYWlyIG1lY2hhbmlzbXMuIE9uZSBtZWNoYW5pc20gcmVjb25uZWN0cyB0aGUgRE5BLCBidXQgb2Z0ZW4gaW4gYSB3YXkgdGhhdCBkaXNhYmxlcyB0aGUgZ2VuZS4=[Qq]
[c]IHN0YXJ0cyBkaXZpZGluZyBvdXQgb2YgY29udHJvbCwgb2Z0ZW4gYmVjb21pbmcgYSB0dW1vciB0aGF0IGNvdWxkIGRldmVsb3AgaW50byBjYW5jZXIu[Qq]
[f]Tm8uIFRoZXJlIGlzIGEgY29ubmVjdGlvbiBiZXR3ZWVuIG11dGF0aW9uIGFuZCBjYW5jZXIsIGJ1dCB0aGVyZSYjODIxNztzIGEgYmV0dGVyIGFuc3dlci4=[Qq]
[q]If scientists add working DNA to a cell where CRISPR-Cas9 is at work, the cell might cut out a defective gene and [hangman] in a working copy of that gene (hint: think of how you cut and ______ words in a word processor).
[c]cGFzdGU=[Qq]
[q]If CRISPR-Cas9 and foreign [hangman] are introduced to a fertilized egg, the result can be a transgenic organism.
[c]RE5B[Qq]
[/qwiz]
3. Understanding CRISPR-Cas9
Restriction enzymes are proteins that have a shape that enables them to bind with and cut up DNA. CRISPR-Cas9 particles consist of a protein/enzyme that’s combined with RNA. The RNA in a CRISPR-Cas9 particle is what binds with the target DNA. The Cas9 particle is what does the cutting.
|
|
|
Like restriction enzymes, CRISPR-Cas9 is an engineered version of a system that evolved in bacteria to fight off viral attacks. In bacteria, CRISPR-Cas-9 acts as a kind of bacterial immune system. The image at left shows a bacteriophage (a) injecting its DNA (c) into a bacterial cell (b). In this representation, the line with colored segments labeled “TracrRNA,” “Cas9,” etc. on the bottom of the diagram represents a small section of a bacterial chromosome.
Enzymes that are associated with the CRISPR system (d) attack the viral DNA, breaking it into fragments (e). One of these fragments (f) will be incorporated into a gene segment called a CRISPR array (j). “G” shows a viral DNA sequence that’s been incorporated into the CRISPR array. The letter “i” shows viral DNA sequences from previous viral attacks. The letter “H” shows DNA segments that separate the viral DNA sequences.
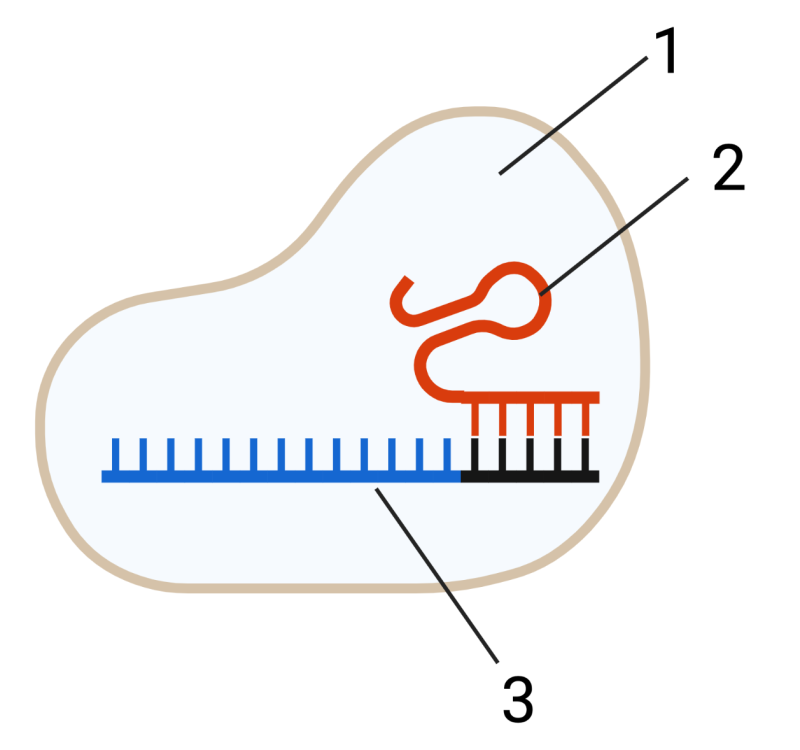
Bacteria use these viral DNA segments to construct CRISPR-Cas9 units. These units (shown in a very simplified form on the right) consist of the Cas9 protein (1) and RNA (2 and 3). The RNA at 2 has a structural function. The RNA at 3 is transcribed from the viral DNA (“g” and “i” above) that was incorporated into the CRISPR array (j, above). That RNA can be thought of as “targeting RNA.”
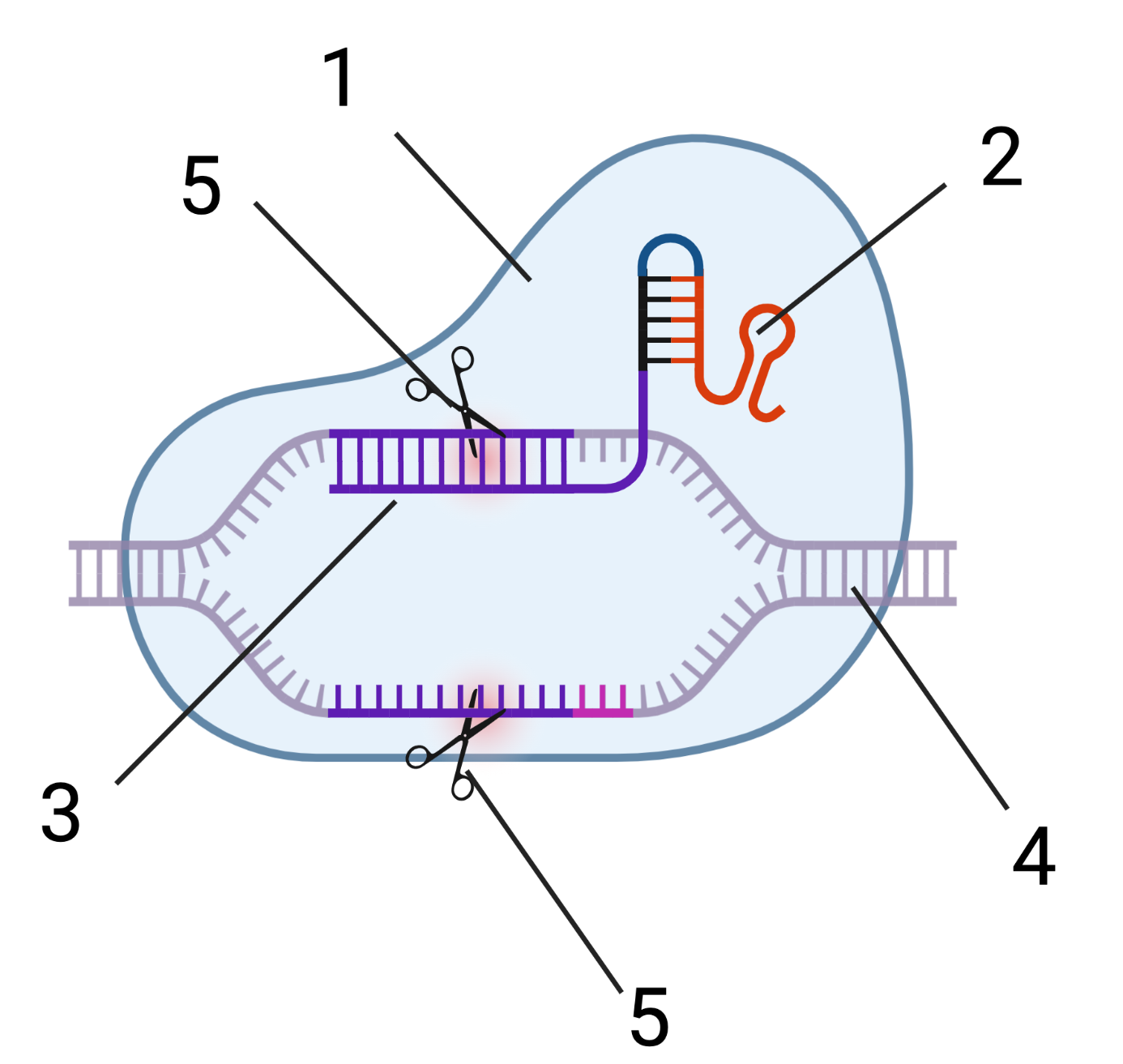
What does this targeting RNA target? It targets the same viral DNA from which it was originally transcribed. When a CRISPR-Cas9 particle encounters viral DNA with a sequence that’s complementary to its targeting RNA (4), an enzymatic portion of Cas9 (5) makes a double-stranded cut in the viral DNA, which short-circuits the viral attack.
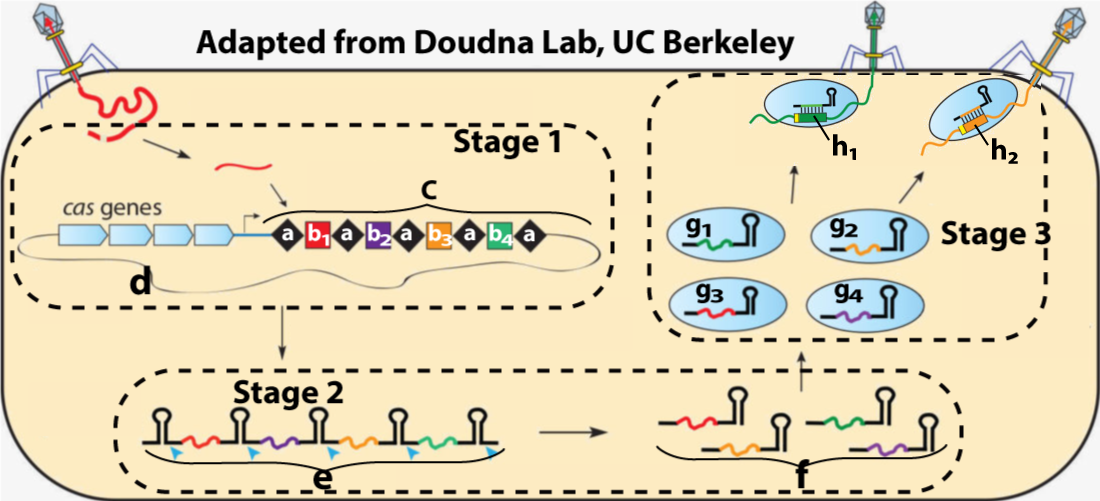 Here’s what the entire system looks like.
Here’s what the entire system looks like.
Stage 1 shows a viral attack and its immediate consequences. Viral DNA from the attack (b1) has been incorporated into the CRISPR array (c) within a bacterial chromosome (d). Viral DNA from previous attacks is represented by b2, b3, and b4.
Stage 2 shows the transcription of CRISPR RNA (e), which then gets cut apart into distinct RNA segments (f). Each of the RNA segments at “f” is complementary to a segment of viral DNA from a previous viral attack.
Stage 3 shows the association of each CRISPR RNA segment with a Cas9 protein. These CRISPR-Cas9 particles will now roam the cytoplasm, “looking” for complementary DNA from bacteriophages that have previously attacked the cell. In the top right of the diagram, you can see CRISPR-Cas9 particles finding such viral RNA (h1 and h2) and binding to it. In the next step (not shown), Cas9 nucleases will cut up this viral RNA, stopping the viral attack.
Understanding the CRISPR-Cas9 system involved a series of discoveries that began in the late 1980s. By 2012, two scientists studying CRISPR — Jennifer Doudna at UC Berkeley and Emmanuele Charpentier, currently at the Max Plank Institute in Berkely — realized that with a bit of engineering, CRISPR-Cas9 could be modified to bind to and cut apart any DNA sequence. All that needed to be done was to create a custom RNA sequence that would complement a targeted DNA sequence.
4. CRISPR-Cas9 as a Gene Editing System
CRISPR-Cas9’s power relates to how easy it makes editing the genes of any organism. Before CRISPR-Cas9, other gene editing systems involved designing proteins that would find specific DNA sequences. Engineering these DNA-binding proteins was both expensive and time-consuming. By contrast, designing custom-made guide RNA for CRISPR is comparatively quick and easy (you can read this 2014 blog post from the Jackson Lab for a comparison of CRISPR with some of the previous techniques).
Paired with sequencing technology, researchers can target any gene of interest by creating a custom sgRNA that seeks out the target sequence and makes a double-stranded cut.
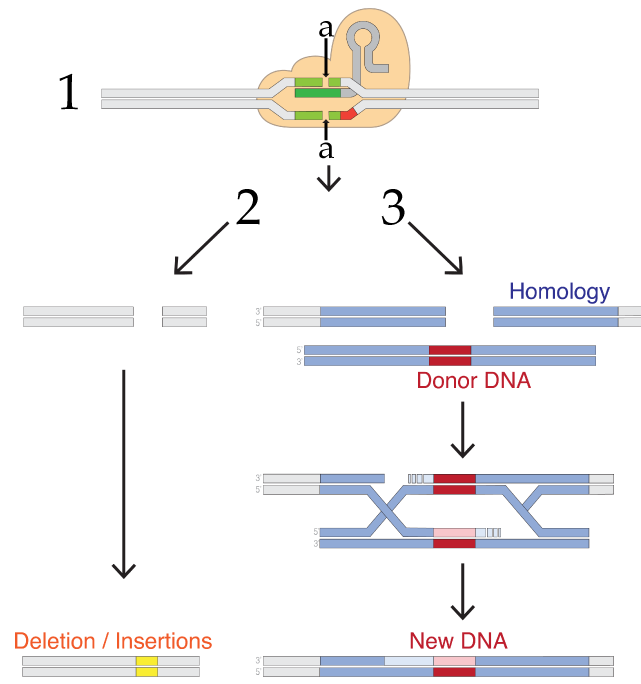
What happens next depends on the researcher’s goals. What follows lists only a few options, but keep in mind that CRISPR technology is changing rapidly.
If the goal is to disable a gene, then you allow what’s shown as process number 2 in the diagram to the right. In any cell, a team of enzymes monitors the DNA for double-stranded breaks, and then works to recombine the broken DNA by reconnecting the broken strands. The type of repair shown on the left is called non-homologous end joining, a term that only makes sense if you compare it to the system on the right side (number 3).
As you learned in unit 5, any sexually reproducing organism inherits one allele from its mother, and the other from its father. If one allele is damaged, then the second allele can be used as a reference to correct the damaged strand. In non-homologous end joining, the homologous template is not used during the DNA repair process. Rather, a team of enzymes grabs the broken ends and trims or adds nucleotides as needed to reconnect them. This process tends to be inaccurate and results in deletions or insertions that wind up disabling the gene.
In homology-directed repair (HDR), the homologous allele is used as a template by DNA repair enzymes, which repair the double-stranded break in a way that leaves the gene in a functional state. Homologous repair opens up the possibility of adding new DNA. In this method, the targeted DNA breakage by CRISPR-Cas9 is followed by the introduction of what’s labeled in figure 7 above as “Donor DNA.” This DNA has ends that are homologous to the broken strand, but with new DNA inserted between the homologous ends. This new DNA could be a working version of a broken gene or a gene from another species. When homologous repair enzymes repair the broken DNA, they’ll wind up inserting this new DNA. This gives genetic engineers a relatively easy way to create recombinant DNA within living cells.
5. Some CRISPR-Cas9 Applications
Gene editing using CRISPR-Cas9 is progressing at such a pace that by the time you read this the applications listed below will have been superseded by newer ones. Searching for “CRISPR Applications” will get you to the latest developments. In the meanwhile, here’s a starting list of some procedures that biologists are working on…
- Potential therapies for inherited genetic disorders. Research programs are underway for using CRISPR to cure or ameliorate the symptoms of genetic disorders such as Duchenne Muscular Dystrophy, Cystic Fibrosis, and Sickle Cell Disease. For an overview of CRISPR-based medicine, read this Web MD Article.
- Fighting Viruses: CRISPR-Cas9’s evolutionary roots are as a counterattack against bacteriophage. Hence, it’s no surprise that the system is being used against human viral pathogens. One application being researched is to target HIV when it’s embedded as a provirus in T-cells. Unlike current therapies, which suppress HIV, this therapy could eradicate it from people who are HIV positive. Similarly, CRISPR is being investigated as a means to eradicate viral DNA from pig organs, making them suitable for transplant into humans (sciencemag.org)
- Agriculture: CRISPR works with plant cells as well as it works in animal cells. CRISPR-based gene editing is already being used to increase pest resistance in crops such as wheat, corn, and tomatoes. You can read more about the use of CRISPR in agriculture here.
- Cancer Research and Therapy
- In some cancers, Killer T cells that would normally attack tumors are deactivated by signals from cancer cells. A planned trial involves using CRISPR-Cas9 to delete two genes in T cells and then infuse them back into the patients, where they’ll be able to seek out and destroy tumors (source: MIT Technology Review).
- The Human Papilloma Virus (HPV) causes genital warts which, if untreated, can progress to cervical cancer. In cervical cancer cell lines (HPV-infected cancer cells that are being grown in cultures), CRISPR-Cas9 targeting of two genes was able to induce these cancerous cells to commit programmed cell suicide, also known as apoptosis. Similar treatments might also be possible for other virally induced cancers, such as Burkitt’s lymphoma, which can be caused by the Epstein Barr Virus (source: Molecular Therapy).
For further Reading about CRISPR and its potential applications (from three of its discoverers), read The Kavli Prize Interview with Jennifer Doudna, Emmanuelle Charpentier, and Virginijus Šikšnys.
6. CRISPR-Cas9: Checking Understanding
[qwiz random = “true” style=”width: 600px !important;” qrecord_id=”sciencemusicvideosMeister1961-Sequencing and CRISPR-Cas9 (v2.0)”] [h]
Sequencing and CRISPR-Cas9
[i]
[q] In the diagram below, a variety of viral DNA fragments are shown at
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IEdvb2QhIEZyYWdtZW50ZWQgdmlyYWwgRE5BIGlzIHNob3duIGF0ICYjODIyMDtlLiYjODIyMTs=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLCBidXQgaGVyZSYjODIxNztzIGEgaGludC4gVGhlIEROQSBpbnNlcnRlZCBieSB0aGUgdmlydXMgaXMgc2hvd24gYXQgJiM4MjIwO2MuJiM4MjIxOyBJZiB0aGF0IEROQSB3ZXJlIGZyYWdtZW50ZWQsIHdoYXQgd291bGQgaXQgbG9vayBsaWtlP8Kg[Qq]
[q] In the diagram below, the viral DNA that will be incorporated into the bacterial chromosome is shown at
[textentry single_char=”true”]
[c]IG Y=[Qq]
[f]IEdvb2QhIFRoZSB2aXJhbCBETkEgZnJhZ21lbnQgdGhhdCB3aWxsIGJlY29tZSBpbmNvcnBvcmF0ZWQgaW50byB0aGUgYmFjdGVyaWFsIGNocm9tb3NvbWUgaXMgYXQgJiM4MjIwO2YuJiM4MjIxO8Kg[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBJbiB0aGUgZGlhZ3JhbSwgZmluZCB0aGUgRE5BIGZyYWdtZW50IHRoYXQmIzgyMTc7cyBnb2luZyB0byBiZSBpbnNlcnRlZCBpbnRvIHRoZSBDUklTUFIgYXJyYXkgKGF0ICYjODIyMDtHJiM4MjIxOykuwqA=[Qq]
[q] In the diagram below, the CRISPR array is shown at
[textentry single_char=”true”]
[c]IG o=[Qq]
[f]IENvcnJlY3QhIFRoZSBDUklTUFIgYXJyYXkgY29uc2lzdHMgb2YgRE5BIHNlcXVlbmNlcyBmcm9tIHByZXZpb3VzIHZpcmFsIGF0dGFja3MgKGcgYW5kIGkpIHNlcGFyYXRlZCBieSBvdGhlciBETkEuwqA=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLCBidXQgaGVyZSYjODIxNztzIGEgaGludC4gVGhlIENSSVNQUiBhcnJheSBjb25zaXN0cyBvZiBETkEgc2VxdWVuY2VzIGZyb20gcHJldmlvdXMgdmlyYWwgYXR0YWNrcyAoZyBhbmQgaSkgc2VwYXJhdGVkIGJ5IG90aGVyIEROQS4=[Qq]
[q] In the diagram below, the CRISPR gene array is found at
[textentry single_char=”true”]
[c]IG M=[Qq]
[f]IEF3ZXNvbWUhIExldHRlciAmIzgyMjA7YyYjODIyMTsgc2hvd3MgdGhlIENSSVNQUiBhcnJheS4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLCBidXQgaGVyZSYjODIxNztzIGEgaGludC4gWW91JiM4MjE3O3JlIGxvb2tpbmcgZm9yIEROQSB0aGF0IGluY29ycG9yYXRlcyBETkEgZnJvbSBwcmV2aW91cyB2aXJhbCBhdHRhY2suwqA=[Qq]
[q] In the diagram below, the bacterial chromosome is indicated by
[textentry single_char=”true”]
[c]IG Q=[Qq]
[f]IEF3ZXNvbWUhIExldHRlciAmIzgyMjA7ZCYjODIyMTsgaW5kaWNhdGVzIHRoZSBiYWN0ZXJpYWwgY2hyb21vc29tZS7CoA==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLCBidXQgaGVyZSYjODIxNztzIGEgaGludC4gVGhlIGJhY3RlcmlhbCBjaHJvbW9zb21lIGlzIGEgY2lyY3VsYXIgcGllY2Ugb2YgRE5BIHRoYXQgaW5jbHVkZXMgdGhlIENSSVNQUiBhcnJheSwgQ1JJU1BSLWFzc29jaWF0ZWQgZ2VuZXMsIGFuZCBhbGwgb2YgdGhlIGJhY3RlcmlhbCBjZWxsJiM4MjE3O3Mgb3RoZXIgZ2VuZXMuwqA=[Qq]
[q] In the diagram below, a connected cluster of CRISPR RNAs is indicated by
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IEF3ZXNvbWUhIExldHRlciAmIzgyMjA7ZSYjODIyMTsgaW5kaWNhdGVzIGEgY29ubmVjdGVkIGNsdXN0ZXIgb2YgQ1JJU1BSIFJOQXM=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLCBidXQgaGVyZSYjODIxNztzIGEgaGludC4gTG9vayBpbiBzdGFnZSAyLiBXaGF0IGNvdWxkIGJlIGRlc2NyaWJlZCBhcyBhICYjODIyMDtjb25uZWN0ZWQgY2x1c3Rlci4mIzgyMjE7[Qq]
[q] In the diagram below, CRISPR RNA that hasn’t yet been associated with the Cas9 protein is shown at
[textentry single_char=”true”]
[c]IG Y=[Qq]
[f]IEF3ZXNvbWUhIExldHRlciAmIzgyMjA7ZiYjODIyMTsgaW5kaWNhdGVzIENSSVNQUiBSTkEuwqA=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLCBidXQgaGVyZSYjODIxNztzIGEgaGludC4gQXQgc3RhZ2UgMywgeW91IGNhbiBzZWUgZm91ciBDUklTUFItQ2FzOXMuIEVhY2ggb2YgdGhlc2UgY29uc2lzdHMgb2YgYSBDYXM5IHByb3RlaW4gKGluIGJsdWUpIGFzc29jaWF0ZWQgd2l0aCBhIENSSVNQUiBSTkEu[Qq]
[q] In the diagram below, viral DNA that’s about the be cut apart by CRISPR-Cas9 is shown at which letter? Note: don’t worry about using a subscript in your answer.
[textentry single_char=”true”]
[c]IG g=[Qq]
[f]IEF3ZXNvbWUhIExldHRlciAmIzgyMjA7aCYjODIyMTsgaW5kaWNhdGVzIGEgdmlyYWwgRE5BIHNlcXVlbmNlIHRoYXQgY29tcGxlbWVudHMgdGhlIENSSVNQUiBSTkEu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLCBidXQgaGVyZSYjODIxNztzIGEgaGludC4gTG9vayBmb3IgdmlyYWwgRE5BIHRoYXQmIzgyMTc7cyBjb21wbGVtZW50YXJ5IHRvIENSSVNQUiBSTkEu[Qq]
[q] In the diagram below, CRISPR’s targeting RNA is indicated by
[textentry single_char=”true”]
[c]IG E=[Qq]
[f]IEF3ZXNvbWUhIExldHRlciAmIzgyMjA7YSYjODIyMTsgaW5kaWNhdGVzIENSSVNQUiYjODIxNztzIHRhcmdldGluZyBSTkEu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLCBidXQgaGVyZSYjODIxNztzIGEgaGludC4gUk5BIGlzIHNpbmdsZS1zdHJhbmRlZC4=[Qq]
[q] In the diagram below, non-homologous end joining is indicated by
[textentry single_char=”true”]
[c]IG Q=[Qq]
[f]IFRlcnJpZmljISBMZXR0ZXIgJiM4MjIwO2QmIzgyMjE7IGluZGljYXRlcyBub24taG9tb2xvZ291cyBlbmQgam9pbmluZy7CoA==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHBhcnQgb2YgdGhlIGRpYWdyYW0gaW4gd2hpY2ggdGhlIGVuZHMgb2YgdGhlIEROQSBhcmUgc3RpdGNoZWQgdG9nZXRoZXIsIHdpdGhvdXQgcmVmZXJlbmNlIHRvIGFueSBvdGhlciBzdHJhbmQgb2YgRE5BLsKg[Qq]
[q] In the diagram below, which letter indicates a process that could be used to deactivate/knock out a gene?
[textentry single_char=”true”]
[c]IG Q=[Qq]
[f]IFRlcnJpZmljISBMZXR0ZXIgJiM4MjIwO2QmIzgyMjE7IGluZGljYXRlcyBub24taG9tb2xvZ291cyBlbmQgam9pbmluZy4gVGhlIG11dGF0aW9ucyB0aGF0IGFyZSBhc3NvY2lhdGVkIHdpdGggdGhhdCBwcm9jZXNzIHVzdWFsbHkgd2luZCB1cCBkZWFjdGl2YXRpbmcgb3Iga25vY2tpbmcgb3V0IGdlbmUgYWN0aXZpdHkuwqA=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHBhcnQgb2YgdGhlIGRpYWdyYW0gaW4gd2hpY2ggdGhlIGVuZHMgb2YgdGhlIEROQSBhcmUgc3RpdGNoZWQgdG9nZXRoZXIsIHdpdGhvdXQgcmVmZXJlbmNlIHRvIGFueSBvdGhlciBzdHJhbmQgb2YgRE5BLsKg[Qq]
[q] In the diagram below, which letter indicates a process that could be used to insert a new gene?
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IFRlcnJpZmljISBMZXR0ZXIgJiM4MjIwO2UmIzgyMjE7IGluZGljYXRlcyBob21vbG9neS1kaXJlY3RlZCByZXBhaXIuIFRoZSBwYXJ0IG9mIHRoZSBzZXF1ZW5jZSBpbmRpY2F0ZWQgYnkgdGhlIHJlZCByZWN0YW5nbGUgaW5kaWNhdGVzIHRoZSBpbnNlcnRpb24gb2YgbmV3IEROQS7CoA==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHBhcnQgb2YgdGhlIGRpYWdyYW0gaW4gd2hpY2ggdGhlIEROQSBpcyByZXBhaXJlZCBieSB1c2luZyBhIGhvbW9sb2dvdXMgdGVtcGxhdGUgYXMgYSByZWZlcmVuY2UuwqA=[Qq]
[/qwiz]
7. Flashcards: Sequencing, and CRISPR
[qdeck bold_text=”false” style=”width: 600px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-Sequencing and CRISPR Flashcards (v2.0)”]
[h] Sequencing and CRISPR
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|16f0034a68510″ question_number=”273″ topic=”6.8.Biotechnology”] What is DNA sequencing?
Illustrative example: Genetic Engineering.
Importance for the AP exam: High
[a] Sequencing is taking a sample of DNA – anything from a small fragment to the entire genome of an organism – and figuring out the specific sequence of A, T, C, and G nucleotides that make it up.
[q]CRISPR evolved from a system used by bacteria to remember previous viral attacks and to fight off new viral attacks. Using the diagram below as a stimulus, explain how that system works.
[a]In response to a viral attack (top left), bacteria will cut up viral DNA into fragments, and incorporate that DNA into a CRISPR array (c) within the bacterial chromosome (d). That DNA will be transcribed into RNA (e), which will be cleaved into separate segments of CRISPR RNA (f). These CRISPR RNAs will then associate with the CAS-9 protein (g1 through g4). Each CRISPR-Cas9 particle will now float in the bacterial cytoplasm, “waiting” to encounter viral DNA with a sequence complementary to the CRISPR RNA. When that happens, the Cas9 particle will act as an nuclease, cutting the viral DNA apart, and preventing viral takeover.
[q]Why is CRISPR such a revolutionary improvement over previous DNA editing tools, such as restriction enzymes
[a]Previous DNA cutting/editing tools, such as restriction enzymes, were proteins. Creating a protein that would bind to a specific DNA sequence was difficult. Because CRISPR-Cas9 finds its target DNA through its guide RNA (3, below), it’s an easily reprogrammable system. If you want to target a specific DNA sequence (4), all that you have to do is to create a complementary RNA sequence (something that’s relatively easy to do with the tools of modern molecular biology).
[q]How can CRISPR-Cas9 be used to insert new genes into an organism?
[a]By modifying its RNA sequence, CRISPR-Cas9 can easily be targeted to snip apart any sequence of DNA. If a piece of DNA is added to a cell where this cut has been made, and if the ends of the DNA has a sequence that matches the flanking ends of the DNA that’s been cut, then the cell will incorporate the new DNA into the broken DNA. This process is called homologous end-joining and can be used to create recombinant DNA
[x][restart]
[/qdeck]
8. Mr. W’s Interview with Jennifer Doudna
In April 2020, I interviewed UC Berkeley Professor Doudna, who won the 2020 Nobel Prize for the discovery of CRISPR, as part of a virtual fundraising luncheon for the Berkeley High School Development Group. Enjoy!
9. What’s Next?
Congratulations! This tutorial ends Topic 6.8 (Genetic Engineering) and Unit 6 (Gene Expression). You’ve completed one of the biggest and most challenging units in AP Bio. To consolidate what you’ve learned, proceed to the Unit 6 Cumulative Review (Objectives, Flashcards, Multiple Choice Questions, and Click-on Challenges).