1. Case Study: Genetic Engineering to Treat Type 1 Diabetes
Genetic engineering is the process of altering the characteristics of an organism by manipulating its genetic material.
Some might argue that humans have been genetically engineering plants and animals for thousands of years through plant and animal breeding. But the kind of genetic engineering we’ll be discussing below is different: it involves direct manipulation of DNA, usually by introducing genes from one species into another.
Genetic engineering has thousands of applications in medicine, agriculture, and industry. In this tutorial, we’ll look at one of genetic engineering’s first applications: synthesizing the hormone insulin to treat Type 1 diabetes.
As you’ll remember from our tutorials about blood sugar homeostasis and diabetes in Unit 4, Type 1 diabetes occurs when people lose the ability to synthesize their own insulin.
Insulin’s role in type 1 diabetes has been understood since 1921. Before that, type 1 diabetes was an untreatable childhood disease. Children who developed type 1 diabetes usually died within a year or two. Once insulin’s role was discovered, doctors quickly figured out that insulin could be extracted from the pancreas of cows and pigs, which produce their own version of insulin that’s almost identical in structure to human insulin.
Once insulin was available, diabetics could monitor their blood glucose levels, and inject insulin as needed. Diabetes shifted from being a death sentence to a treatable condition. (If you want to learn more about the history of insulin’s discovery, you can do so in this article from the Science History Institute or this one from the American Diabetes Association.)
Insulin from cattle and pigs was a lifesaver. But it wasn’t perfect. The tiny differences between animal and human insulin sometimes led to allergic reactions. And that’s where genetic engineering comes in.
2. Through Genetic Engineering, Bacteria can be Modified to Produced Human Insulin
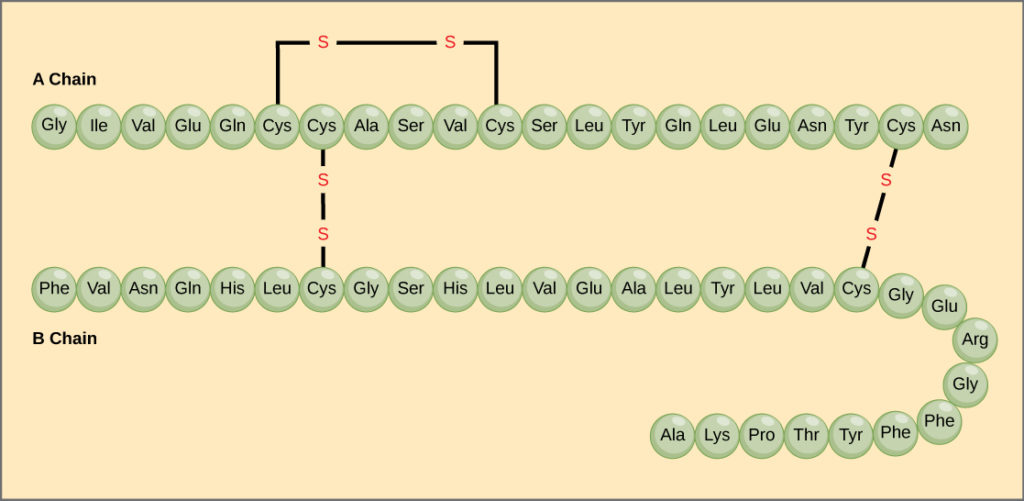
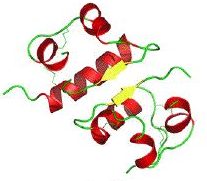
Insulin is a protein that consists of 51 amino acids. Its biologically active form has a quaternary structure consisting of two polypeptide chains linked by two disulfide bridges. A third disulfide bridge within the alpha chain causes a hairpin turn, which you can see in the cartoon structure on the right in the images below.
 |
 |
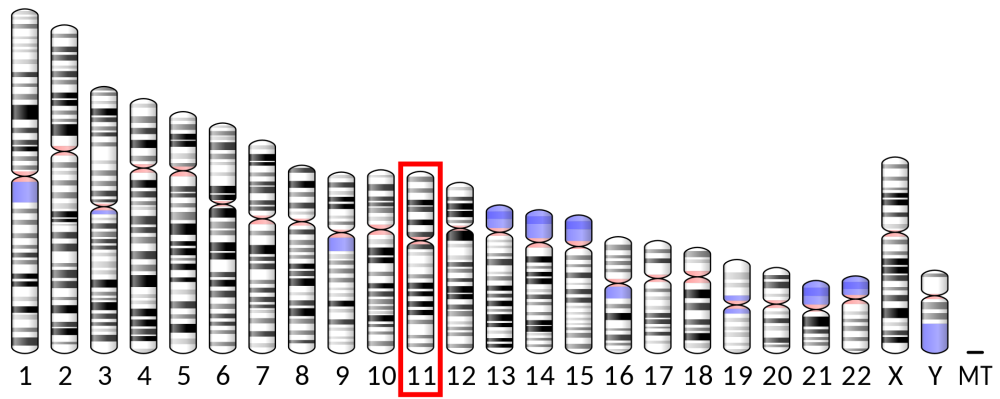
In humans, the gene for insulin is found on chromosome 11. Through the techniques of genetic engineering, that gene can be inserted into a bacterial plasmid. Insertion of that plasmid into a bacterium such as E. coli creates a genetically engineered organism that has a new phenotype: it will secrete human insulin, which can then be used to treat diabetes.
Here are the details.
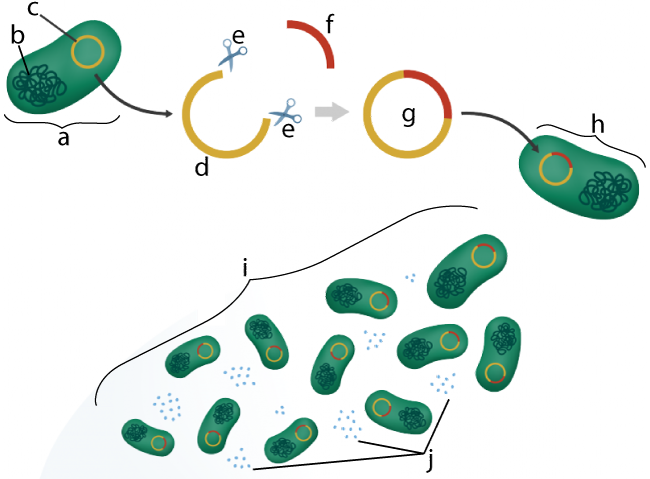
In the diagram to the right, “a” is an E. coli bacterial cell. E. coli is a bacterial species often used in genetic engineering.
In addition to E. coli’s main chromosome (“b”) it also has a plasmid (shown at “c”), a small loop of DNA that we discussed in a previous tutorial in AP Bio Unit 6. If you need to review the role of plasmids in bacterial conjugation and transformation, click the previous link.
Using techniques of genetic engineering (some of which we’ll expand upon below), a plasmid can be extracted from a bacterial cell, and then, using enzymes (e) that act as a kind of molecular scissors, the plasmid can be cut open (as shown at “d”).
This plasmid DNA can then be combined with DNA from another species, such as the human insulin gene, represented by “f”. This creates a piece of recombinant DNA (g). Recombinant DNA is DNA that is a combination of DNA from two sources. In this case, it’s a plasmid that contains both bacterial DNA and the human insulin gene. Then, through engineered bacterial transformation, the genetically engineered plasmid is re-inserted into a bacterial cell (h). Note that we learned about transformation in our tutorial about horizontal gene transfer in the last tutorial.
Once inside its bacterial host, this recombinant DNA will function like any other type of DNA. Every time the bacterial cell reproduces, it will copy the recombinant plasmid (i). The recombinant plasmid’s DNA will also be transcribed and translated, producing human insulin (j). Additional techniques will collect the insulin. Once distributed to diabetics, this genetically engineered insulin can be used by diabetics to control their diabetes.
3. Checking Understanding: Genetically Engineered Insulin
[qwiz random = “true” qrecord_id=”sciencemusicvideosmeister1961-Genetic Engineering Overview (v2.0)”] [h]
Genetic Engineering Overview
[i]
[q]Genetically engineered insulin is used to treat [hangman].
[c]ZGlhYmV0ZXM=[Qq]
[q] In the diagram below, which letter represents the main bacterial chromosome?
[textentry single_char=”true”]
[c]IG I=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIG1haW4gYmFjdGVyaWFsIGNocm9tb3NvbWUgaXMgYXQgJiM4MjIwO2IuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgbWFpbiBiYWN0ZXJpYWwgY2hyb21vc29tZSBpcyB0aGUgbGFyZ2VzdCBwaWVjZSBvZiBETkEgdGhhdCB5b3UgY291bGQgZmluZCBpbiB0aGUgY2VsbC7CoA==[Qq]
[q] In the diagram below, which letter represents an unmodified plasmid?
[textentry single_char=”true”]
[c]IG M=[Qq]
[f]IE5pY2Ugam9iLsKgICYjODIyMDtDJiM4MjIxOyByZXByZXNlbnRzIGFuIHVubW9kaWZpZWQgcGxhc21pZC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLg==[Qq]
[q] In the diagram below, which letter represents the human insulin gene?
[textentry single_char=”true”]
[c]IG Y=[Qq]
[f]IFllcy4gVGhlIGh1bWFuIGluc3VsaW4gZ2VuZSBpcyBhdCAmIzgyMjA7Zi4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3QuIEhlcmUmIzgyMTc7cyBhIGhpbnQuIEluIHRoaXMgdHlwZSBvZiBnZW5ldGljIGVuZ2luZWVyaW5nLCB0aGUgZ2VuZSBmb3IgaHVtYW4gaW5zdWxpbiBpcyBhZGRlZCB0byB0aGUgYmFjdGVyaWFsIHBsYXNtaWQuIFdoYXQmIzgyMTc7cyB0aGUgb25seSB0aGluZyB0aGF0IGNvdWxkIHJlcHJlc2VudCB0aGlzIGFkZGl0aW9uYWwgRE5BPw==[Qq]
[q] In the diagram below, a plasmid with recombinant DNA is shown at
[textentry single_char=”true”]
[c]IG c=[Qq]
[f]IE5pY2Ugam9iISBMZXR0ZXIgJiM4MjIwO2cmIzgyMjE7IHJlcHJlc2VudHMgYSByZWNvbWJpbmFudCBwbGFzbWlkLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJZiBEIHJlcHJlc2VudHMgdGhlIGJhY3RlcmlhbCBwbGFzbWlkLCBhbmQgRiByZXByZXNlbnRzIHRoZSBodW1hbiBpbnN1bGluIGdlbmUsIHdoYXQgaXMgdGhlIG9ubHkgbGV0dGVyIHRoYXQgY291bGQgcmVwcmVzZW50IGEgcmVjb21iaW5hbnQgcGxhc21pZD8=[Qq]
[q] In the diagram below, which letter indicates the reproduction of genetically engineered offspring?
[textentry single_char=”true”]
[c]IG k=[Qq]
[f]IFllcyEgVGhlIGxldHRlciAmIzgyMjA7aSYjODIyMTsgc2hvd3MgdGhlIG9mZnNwcmluZyBvZiB0aGUgZ2VuZXRpY2FsbHkgZW5naW5lZXJlZCBwYXJlbnQgY2VsbC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIFdoaWNoIGxldHRlciBzaG93cyB0aGUgb2Zmc3ByaW5nIG9mIHRoZSBnZW5ldGljYWxseSBlbmdpbmVlcmVkIHBhcmVudCBjZWxsPw==[Qq]
[q] In the diagram below, which letter indicates the production of genetically engineered insulin?
[textentry single_char=”true”]
[c]IG o=[Qq]
[f]IFllcyEgTGV0dGVyICYjODIyMDtqJiM4MjIxOyBpbmRpY2F0ZXMgdGhlIHByb2R1Y3Rpb24gb2YgZ2VuZXRpY2FsbHkgZW5naW5lZXJlZCBpbnN1bGluLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIEluc3VsaW4gd2lsbCBiZSBzZWNyZXRlZCBmcm9tIHRoZSBnZW5ldGljYWxseSBlbmdpbmVlcmVkIGNlbGxzLCBhZnRlciB3aGljaCBpdCBjYW4gYmUgaGFydmVzdGVkIGZvciBkaXN0cmlidXRpb24uIFdoaWNoIGxldHRlciBzaG93cyBzb21ldGhpbmcgdGhhdCYjODIxNztzIGJlZW4gc2VjcmV0ZWQgZnJvbSBhIGNlbGw/[Qq]
[q]DNA that is a combination of DNA from two different sources is called [hangman] DNA.
[c]cmVjb21iaW5hbnQ=[Qq]
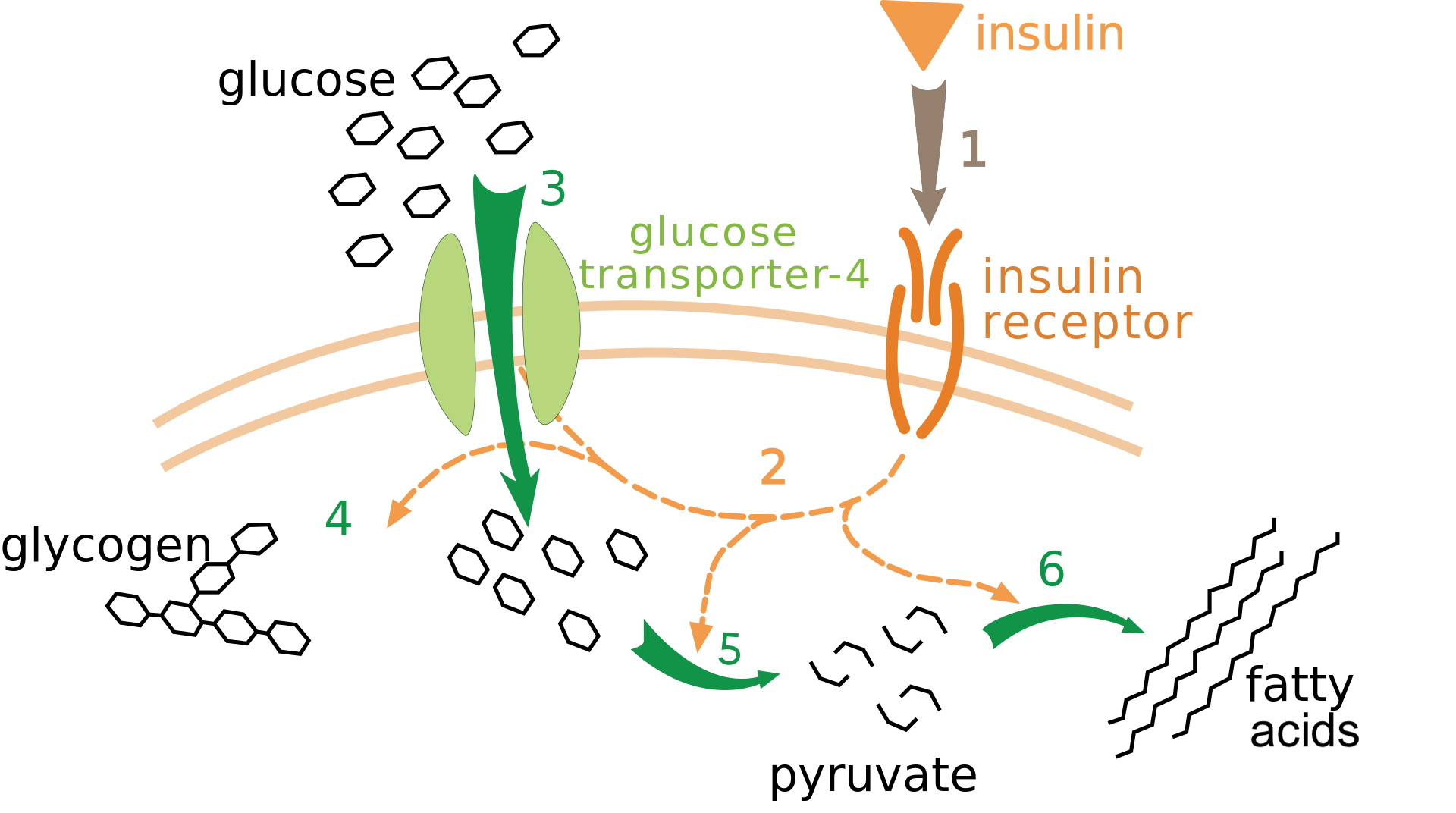
[q]In the diagram below, letter “b” represents [hangman]. Number 3 shows glucose entering into the cell by a type of membrane transport called [hangman] [hangman].
[c]aW5zdWxpbg==[Qq]
[c]ZmFjaWxpdGF0ZWQ=[Qq]
[c]ZGlmZnVzaW9u[Qq]
[q]Insulin is a [hangman] hormone, the function of which is to reduce the amount of [hangman] in the bloodstream.
[c]cHJvdGVpbg==[Qq]
[c]Z2x1Y29zZQ==[Qq]
[/qwiz]
4. Making Recombinant DNA
The techniques of genetic engineering were first developed in the early 1970s. The first breakthrough came about when Paul Berg, working at Stanford University, created the first recombinant DNA molecules. Berg, along with Walter Gilbert and Fredrick Sanger, won the 1980 Nobel Prize for this work.
 To make recombinant DNA, you have to be able to cut apart DNA from at least two different sources and combine them. In the big-picture view of genetic engineering above, we referred to enzymes that act as “molecular scissors.” These enzymes are called restriction endonucleases, or restriction enzymes.
To make recombinant DNA, you have to be able to cut apart DNA from at least two different sources and combine them. In the big-picture view of genetic engineering above, we referred to enzymes that act as “molecular scissors.” These enzymes are called restriction endonucleases, or restriction enzymes.
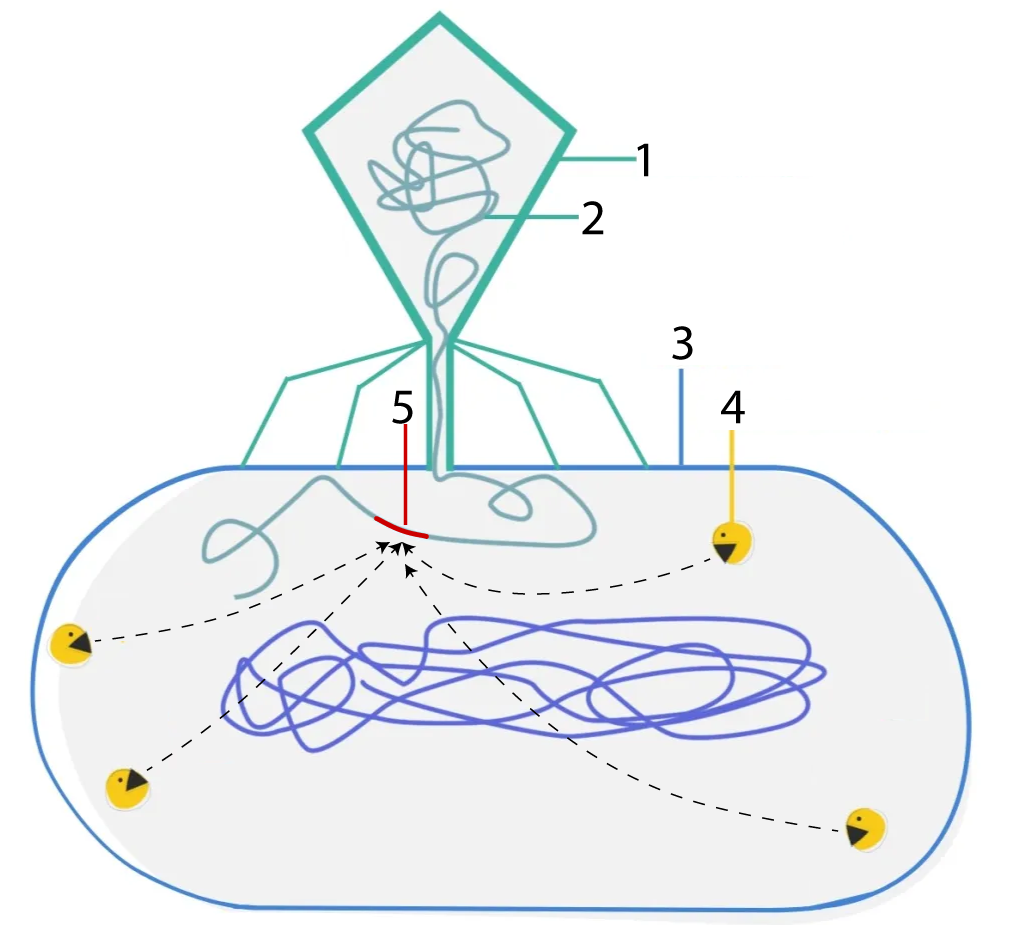
Like many of the molecular tools used by genetic engineers, restriction enzymes first evolved in bacteria. In nature, these enzymes act as a defense against attacks from viruses, and they work by cutting up viral DNA. As we previously learned in our tutorial about the lytic cycle, bacterial viruses (or bacteriophages) start their attack by landing on bacterial cells and then injecting their DNA into the cell. In the diagram to the right, you can see a bacteriophage is shown at 1, and its DNA at 2.
After the phage DNA is injected, the bacteria launch a counterattack. Their restriction enzymes (the PacMan-like structures at “4”) can identify specific sequences in viral DNA (indicated by the red sequence at 5), and snip the DNA apart at that point. Cutting up the viral DNA keeps viruses from taking over the bacterial cells into which they’ve injected their DNA. In other words, these restriction enzymes restricted the ability of bacterial viruses to take over bacterial cells: hence, “restriction enzymes.” The term endonuclease refers to the fact that these enzymes make a cut within a segment of DNA. Both terms — restriction enzyme and restriction endonuclease — are commonly used.
Here’s a closeup view of how one restriction endonuclease works.
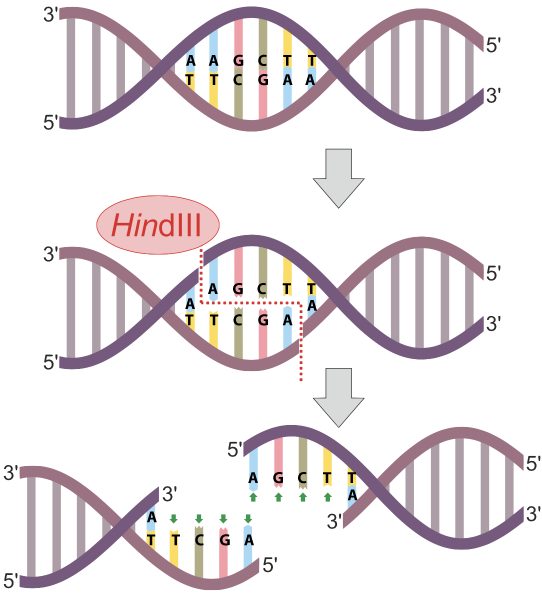
In the example at left, the restriction endonuclease HindIII recognizes the nucleotide sequence AAGCTT and cuts the DNA between the two As. Note that because the sequence is a palindrome (it’s the same in the 5′ to 3′ direction on both strands), the cut winds up being double-stranded, and completely severs the DNA.
Since the time of their initial discovery, over 4000 restriction enzymes have been identified. If you want to learn more about them, follow this link, which will open in a new tab.
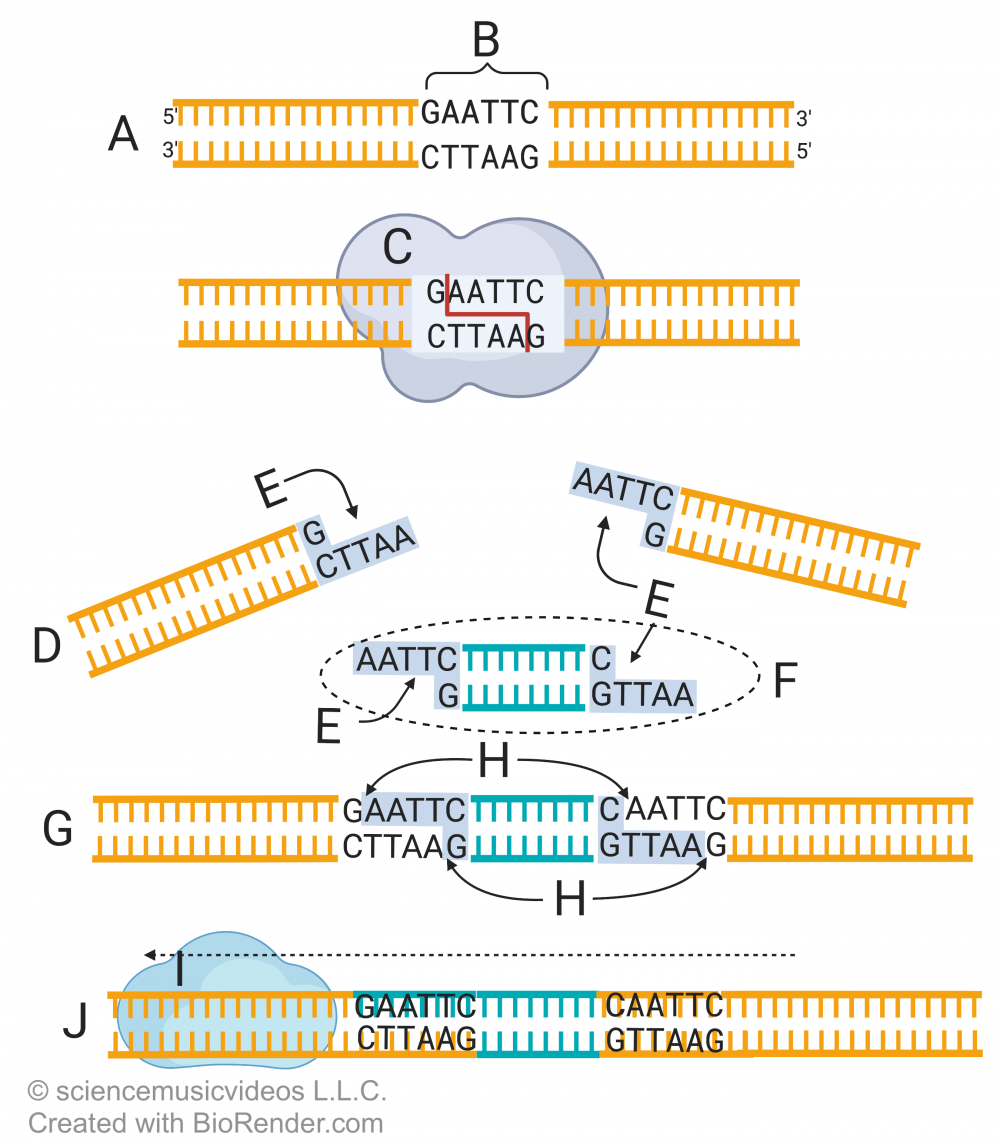
Here’s how restriction enzymes are used to create recombinant DNA. Letter A shows a stretch of DNA, within which is the sequence GAATTC. That sequence is a restriction site (B). It’s recognized by a restriction enzyme (C), which cuts the sugar-phosphate bonds in the DNA between the nucleotide bases guanine and adenine (just look for the arrow between the G and A in the DNA).
The result is two DNA fragments (D), each with a “sticky end” (E). The ends are sticky because they contain exposed nucleotide bases that are ready to form hydrogen bonds with complementary nucleotides.
Letter “F” represents a second stretch of DNA from another species or organism that’s been cut with the same restriction enzyme. If that piece of DNA (F) is mixed with the DNA fragments at “D,” then the two can combine as is shown in “G.” At this point, the DNA is held together just by the hydrogen bonds between the sticky ends of each fragment. “H” represents a gap: a missing sugar-phosphate bond that would connect the fragments into a single strand.
The last step involves a second enzyme, DNA ligase (i). DNA ligase creates sugar-phosphate bonds between adjacent nucleotides. Once ligase has done its work, you have recombinant DNA (J): DNA molecule with sequences from different species.
5. Getting the DNA for Human Insulin
Shortly after Berg created the first recombinant DNA molecules, Herbert Boyer and Stanley Cohen introduced a recombinant plasmid into a bacterial cell: they took a gene for resistance to kanamycin (an antibiotic) and inserted it into a plasmid. When they then inserted this modified plasmid into bacterial cells, the resulting bacteria were kanamycin resistant. In 1973, they introduced a gene from a toad into bacteria, proving that genes could be moved from animals into bacteria.
Once these techniques were developed, scientists (and investors) began to think about how they could profit from this technology. The huge market for insulin (over a million type-1 diabetics who need daily insulin injections) made the genetic engineering of human insulin an early target of those efforts, and that target was pursued by two nascent biotechnology firms in the early 1980s: Biogen and Genentech. The goal became the successful insertion of the genes for human insulin into a bacterial plasmid.
But how was that human DNA to be acquired? Two techniques were used.
The first, pursued by a biotech company called Genentech, involved creating an artificial copy of the insulin gene. Knowing the primary structure of the human insulin protein (see above) scientists could work backward to construct DNA that would code for that protein.
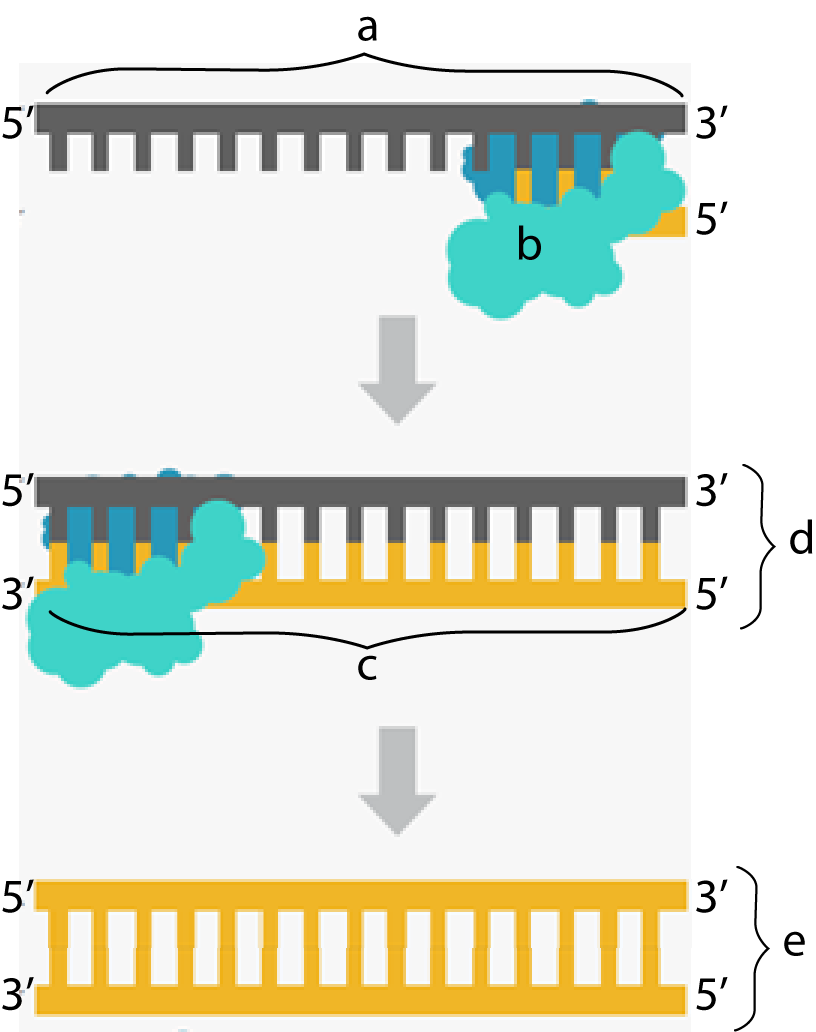
The second way the insulin gene was isolated was used by the biotech company Biogen. Biogen deployed a biochemical trick that evolved in a class of viruses called retroviruses (such as HIV), which we studied earlier in this unit. Retroviruses store their genetic information in the nucleic acid RNA and then use an enzyme called reverse transcriptase to convert their RNA into DNA. Taking a page from this viral playbook, Biogen did the following:
- Cells that produced insulin were isolated from pancreatic tissue and grown in tissue culture.
- The messenger RNA from these cells was isolated, and then reverse transcriptase was used to create DNA that complemented the insulin-coding RNA. In the diagram above and to the right, “a” is mRNA, and “b” is reverse transcriptase.
- As reverse transcriptase moves along the RNA, it lays down new deoxyribonucleotides, synthesizing a new strand in the 5′ to 3′ direction. This creates a strand that’s half RNA and half DNA (shown at “d”).
- Subsequent manipulation with additional enzymes removes the RNA, replacing it with DNA. The result is shown at “e.”
DNA made this way is called cDNA, for complementary DNA. Unlike DNA that would be spliced out of a eukaryotic chromosome, it has the advantage of being intron-free, and only contains the exons for the insulin protein.
Either DNA would have worked. Genentech wound up being the winner. Genentech sold the licensing rights to genetically engineered insulin to Eli Lilly, the U.S.’s biggest producer of insulin.
6. Genetic Engineering and Recombinant DNA: Interactive Diagrams
[qwiz qrecord_id=”sciencemusicvideosmeister1961-Genetic Engineering and Recombinant DNA Diagrams (v2.0)”]
[h]Genetic Engineering and Recombinant DNA
[i]
[q labels = “top”]
[l]human DNA
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]unmodified plasmid
[fx] No. Please try again.
[f*] Great!
[l]recombinant DNA
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l]genetically modified bacterium
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[q labels = “top”]
[l]Ligase
[fx] No. Please try again.
[f*] Good!
[l]recombinant DNA
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]restriction enzyme
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]sticky ends
[fx] No. Please try again.
[f*] Excellent!
[q labels = “top”]
[l]complementary DNA
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]hybrid RNA/DNA
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]reverse transcriptase
[fx] No. Please try again.
[f*] Good!
[q labels = “top”]
[l]bacterial cell wall
[fx] No. Please try again.
[f*] Great!
[l]capsid
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]phage DNA
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l]restriction enzyme
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]restriction site
[fx] No. Please try again.
[f*] Good!
[/qwiz]
7. The Genie’s out of the Bottle
Watson and Crick described the structure of DNA in 1953. By 1973, DNA was being engineered to suit human ends. Now, using techniques very similar to those used to develop genetically engineered insulin, recombinant DNA is everywhere. It’s being deployed to create drugs to treat many diseases, including AIDS, a variety of cancers, cystic fibrosis, hemophilia, genital warts, hepatitis, and human growth hormone deficiency.
The 1980s also saw the development of the first transgenic animals: animals whose genomes were altered through the insertion of genes from other species. These include
- mice with human genes (widely used for research purposes),
- transgenic goats which produce milk that contains substances that prevent the formation of harmful blood clots during surgery (the drugs are extracted from the milk);
- pigs modified with the goal that their organs can be transplanted into humans;
Transgenic crops (also referred to as genetically modified crops or GMOs) have been widely in use since the 1990s. It’s estimated that in 2016, 10% of the world’s cropland was planted with genetically modified plants. (Wikipedia). These include crops such as soybeans, maize (corn), and cotton, which have been modified for increased resistance to pests, and reduction of spoilage.
Transgenic crops can also have improved nutrient profiles. An example of the latter is Golden Rice (Wikipedia): rice that’s been genetically modified to produce beta carotene. Beta carotene is a precursor to vitamin A. Vitamin A deficiency causes hundreds of thousands of childhood deaths in regions where people consume inadequate amounts of that vitamin in their diets. In late 2019, the government of the Philippines approved Golden Rice for human consumption, making it the first Asian country to approve Golden Rice for direct use.
That’s not to say that genetically modified crops are without their critics. For an overview of the controversies related to genetically modified organisms and GMO food regulation, read this on Wikipedia,
8. Checking Understanding: Genetically Engineered Insulin
The following quiz will help you master everything you’ve read above.
[qwiz random = “true” qrecord_id=”sciencemusicvideosmeister1961-Genetic Engineering Quiz (v2.0)”] [h]
Genetic Engineering Quiz
[i]
[q] In the diagram below, a eukaryotic gene of interest is indicated by
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IFllcy4gMSBpcyB0aGUgZ2VuZSBvZiBpbnRlcmVzdC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIFRoZSBETkEgaXMgZ29pbmcgdG8gYmUgY3V0IHdpdGggYSByZXN0cmljdGlvbiBlbnp5bWUgdG8gaXNvbGF0ZSB0aGUgZ2VuZS4gSWYgcmVzdHJpY3Rpb24gZW56eW1lcyBhcmUgYXQgJiM4MjIwOzIsJiM4MjIxOyB0aGVuIHdoaWNoIG51bWJlciBoYXMgdG8gYmUgdGhlIGV1a2FyeW90aWMgZ2VuZSBvZiBpbnRlcmVzdD8=[Qq]
[q] In the diagram below, restriction enzymes are at which number?
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]TmljZSBqb2IhIFJlc3RyaWN0aW9uIGVuenltZXMgYXJlIHJlcHJlc2VudGVkIGJ5IDIu[Qq]
[c]ZW50ZXIgd29yZA==[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]Tm8uIFJlc3RyaWN0aW9uIGVuenltZXMgYXJlIHVzZWQgdG8gY3V0IEROQS4gV2hhdCYjODIxNztzIHRoZSBvbmx5IHRoaW5nIGluIHRoaXMgZGlhZ3JhbSB0aGF0IGNvdWxkIGluZGljYXRlIHNvbWV0aGluZyB0aGF0IGN1dHMgc29tZXRoaW5nIGVsc2Ugb3Blbj8=[Qq]
[q] In the diagram below, a non-recombinant plasmid is indicated by
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]V2F5IHRvIGdvLiBUaGUgdW5tb2RpZmllZCBwbGFzbWlkIGlzIGluZGljYXRlZCBieSAmIzgyMjA7My4mIzgyMjE7[Qq]
[c]ZW50ZXIgd29yZA==[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQ6IGEgcGxhc21pZCBpcyBhIGNpcmNsZSBvZiBleHRyYS1jaHJvbW9zb21hbCBiYWN0ZXJpYWwgRE5BLiBGaW5kIHRoZSBjaXJjbGUmIzgyMzA7[Qq]
[q] In the diagram below, DNA ligase is indicated by
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IFllcyEgTGlnYXNlIGlzIGF0IG51bWJlciA0Lg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMaWdhc2UmIzgyMTc7cyBmdW5jdGlvbiBpcyB0byBjcmVhdGUgc3VnYXItcGhvc3BoYXRlIGJvbmRzIGJldHdlZW4gRE5BIGZyYWdtZW50cy4gV2hhdCYjODIxNztzIG9uIHRoaXMgZGlhZ3JhbSB0aGF0IGNvdWxkIHJlcHJlc2VudCB0aGF0P8Kg[Qq]
[q] In the diagram below, fragments of DNA with “sticky ends” are represented by
[textentry single_char=”true”]
[c]ID U=[Qq]
[f]IEF3ZXNvbWUhIFRoZSBzdGlja3kgZW5kcyBhcmUgcmVwcmVzZW50ZWQgYnkgJiM4MjIwOzUuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBTdGlja3kgZW5kcyBhcmUgZXhwb3NlZCBiYXNlcyB0aGF0IHdpbGwgZm9ybSBoeWRyb2dlbiBib25kcyB3aXRoIG9uZSBhbm90aGVyLiBXaGVuIHRoZSBzYW1lIHJlc3RyaWN0aW9uIGVuenltZSBpcyB1c2VkIHRvIGN1dCBETkEsIHRoZSByZXN1bHRzIHdpbGwgYmUgZnJhZ21lbnRzIHdpdGggdGhlc2Ugc3RpY2t5IGVuZHMu[Qq]
[q] In the diagram below, a recombinant plasmid is shown at
[textentry single_char=”true”]
[c]ID Y=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIHJlY29tYmluYW50IHBsYXNtaWQgaXMgYXQgJiM4MjIwOzYuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBUaGUgcmVjb21iaW5hbnQgcGxhc21pZCB3aWxsIGNvbnNpc3Qgb2YgYSBodW1hbiBnZW5lICgxKSwgYW5kIHRoZSBvcmlnaW5hbCBwbGFzbWlkLg==[Qq]
[q] Organisms that have been modified to contain DNA from more than one species are called[hangman]. Often, these organisms are referred to as genetically [hangman] organisms, or GMOs.
[c]IHRyYW5zZ2VuaWM=[Qq]
[f]IEdvb2Qh[Qq]
[c]IG1vZGlmaWVk[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] Enzymes used to cut DNA are called[hangman] enzymes.
[c]IHJlc3RyaWN0aW9u[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] DNA [hangman]is the enzyme that connects DNA fragments by creating sugar-phosphate bonds.
[c]IGxpZ2FzZQ==[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] [hangman] DNA is DNA that has been pieced together from multiple sources.
[c]IFJlY29tYmluYW50[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] In the diagram below, a restriction site is shown at letter
[textentry single_char=”true”]
[c]IE I=[Qq]
[f]IEdyZWF0ISAmIzgyMjA7QiYjODIyMTsgc2hvd3MgYSByZXN0cmljdGlvbiBzaXRlLsKg[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgcmVzdHJpY3Rpb24gc2l0ZSBpcyB3aGVyZSB0aGUgcmVzdHJpY3Rpb24gZW56eW1lcyB3aWxsIGN1dCBhcGFydCB0aGUgRE5BLg==[Qq]
[q] In the diagram below, a restriction enzyme is shown at
[textentry single_char=”true”]
[c]IG M=[Qq]
[f]IEV4Y2VsbGVudCEgJiM4MjIwO0MmIzgyMjE7IHNob3dzIGEgcmVzdHJpY3Rpb24gZW56eW1lLsKg[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgcmVzdHJpY3Rpb24gZW56eW1lIGlzIHdoYXQgY3V0cyB0aGUgRE5BIGludG8gZnJhZ21lbnRzLg==[Qq]
[q] In the diagram below, “sticky ends” are shown at
[textentry single_char=”true”]
[c]IE U=[Qq]
[f]IENvcnJlY3QhIFN0aWNreSBlbmRzIGFyZSBzaG93biBhdCAmIzgyMjA7RS4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgJiM4MjIwO3N0aWNreSBlbmRzJiM4MjIxOyBhcmUgZXhwb3NlZCBiYXNlcywgcmVhZHkgdG8gZm9ybSBoeWRyb2dlbiBib25kcyB3aXRoIGZyYWdtZW50cyB0aGF0IGhhdmUgY29tcGxlbWVudGFyeSBzZXF1ZW5jZXMu[Qq]
[q] In the diagram below, gaps that need to be sealed together by DNA ligase are shown at
[textentry single_char=”true”]
[c]IE g=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtIJiM4MjIxOyByZXByZXNlbnRzIHRoZSBnYXBzIChhIHN1Z2FyLXBob3NwaGF0ZSBib25kIHRoYXQgbmVlZHMgdG8gYmUgY2F0YWx5emVkKS7CoA==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgZ2FwcyBhcmUgd2hhdCBnZXQgc2VhbGVkIGJ5IEROQSBsaWdhc2UgKHNob3duIGF0ICYjODIyMDtJJiM4MjIxOyku[Qq]
[q] In the diagram below, DNA ligase is shown at
[textentry single_char=”true”]
[c]IE k=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtJJiM4MjIxOyByZXByZXNlbnRzIGxpZ2FzZS7CoA==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBMaWdhc2Ugc2VhbHMgdGhlIGdhcHMgdGhhdCBhcmUgc2hvd24gYXQgJiM4MjIwO0guJiM4MjIxOw==[Qq]
[q] In the diagram below, completed recombinant DNA is shown at
[textentry single_char=”true”]
[c]IE o=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtKJiM4MjIxOyBpcyB0aGUgZmluaXNoZWQgcmVjb21iaW5hbnQgRE5BLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBSZWNvbWJpbmFudCBETkEgY29tYmluZXMgdGhlIGZyYWdtZW50cyBhdCAmIzgyMjA7RCYjODIyMTsgYW5kICYjODIyMDtGLiYjODIyMTs=[Qq]
[q] DNA made from RNA using reverse transcriptase is known as [hangman] DNA.
[c]IGNvbXBsZW1lbnRhcnk=[Qq]
[f]IEdyZWF0IQ==[Qq]
[q multiple_choice=”true”] The enzyme responsible for creating sugar-phosphate bonds between DNA fragments is/are
[c]IEROQSBwb2x5bWVyYXNl[Qq]
[f]IE5vLiBETkEgcG9seW1lcmFzZSBpcyBhIGtleSBlbnp5bWUgcmVzcG9uc2libGUgZm9yIHN5bnRoZXNpemluZyBuZXcgRE5BLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBjcmVhdGluZyB0aGVzZSBzdWdhci1waG9zcGhhdGUgYm9uZHMgaXMgY2FsbGVkICYjODIyMDtsaWdhdGlvbi4mIzgyMjE7[Qq]
[c]IFJldmVyc2UgdHJhbnNjcmlwdGFzZQ==[Qq]
[f]IE5vLiBSZXZlcnNlIHRyYW5zY3JpcHRhc2UgdGFrZXMgUk5BIGFuZCByZXZlcnNlIHRyYW5zY3JpYmVzIGl0IGludG8gRE5BLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBjcmVhdGluZyB0aGVzZSBzdWdhci1waG9zcGhhdGUgYm9uZHMgaXMgY2FsbGVkICYjODIyMDtsaWdhdGlvbi4mIzgyMjE7[Qq]
[c]IFJlc3RyaWN0aW9uIGVuenltZXM=[Qq]
[f]IE5vLiBSZXN0cmljdGlvbiBlbnp5bWVzIGFyZSByZXNwb25zaWJsZSBmb3IgY3V0dGluZyBETkEgaW50byBmcmFnbWVudHMuIEhlcmUmIzgyMTc7cyBhIGhpbnQ6IGNyZWF0aW5nIHRoZXNlIHN1Z2FyLXBob3NwaGF0ZSBib25kcyBpcyBjYWxsZWQgJiM4MjIwO2xpZ2F0aW9uLiYjODIyMTs=[Qq]
[c]IEROQSBs aWdhc2U=[Qq]
[f]IENvcnJlY3QhIEROQSBMaWdhc2UgY3JlYXRlcyBzdWdhci1waG9zcGhhdGUgYm9uZHMgYmV0d2VlbiBhZGphY2VudCBmcmFnbWVudHMgb2YgRE5BLg==[Qq]
[q multiple_choice=”true”] The enzyme that can synthesize DNA from an RNA template is
[c]IEROQSBwb2x5bWVyYXNl[Qq]
[f]IE5vLiBETkEgcG9seW1lcmFzZSBpcyBhIGtleSBlbnp5bWUgcmVzcG9uc2libGUgZm9yIHN5bnRoZXNpemluZyBuZXcgRE5BLg==[Qq]
[c]IFJldmVyc2UgdHJh bnNjcmlwdGFzZQ==[Qq]
[f]IE5pY2Ugam9iLiBSZXZlcnNlIHRyYW5zY3JpcHRhc2UgdGFrZXMgUk5BIGFuZCByZXZlcnNlIHRyYW5zY3JpYmVzIGl0IGludG8gRE5BLg==[Qq]
[c]IFJlc3RyaWN0aW9uIGVuenltZXM=[Qq]
[f]IE5vLiBSZXN0cmljdGlvbiBlbnp5bWVzIGFyZSByZXNwb25zaWJsZSBmb3IgY3V0dGluZyBETkEgaW50byBmcmFnbWVudHM=[Qq]
[c]IEROQSBsaWdhc2U=[Qq]
[f]IE5vLiBETkEgTGlnYXNlIGNyZWF0ZXMgc3VnYXItcGhvc3BoYXRlIGJvbmRzIGJldHdlZW4gYWRqYWNlbnQgZnJhZ21lbnRzIG9mIEROQS4=[Qq]
[q multiple_choice=”true”] The enzyme that can cut DNA into fragments is
[c]IEROQSBwb2x5bWVyYXNl[Qq]
[f]IE5vLiBETkEgcG9seW1lcmFzZSBpcyBhIGtleSBlbnp5bWUgcmVzcG9uc2libGUgZm9yIHN5bnRoZXNpemluZyBuZXcgRE5BLg==[Qq]
[c]IFJldmVyc2UgdHJhbnNjcmlwdGFzZQ==[Qq]
[f]IE5vLiBSZXZlcnNlIHRyYW5zY3JpcHRhc2UgdGFrZXMgUk5BIGFuZCByZXZlcnNlIHRyYW5zY3JpYmVzIGl0IGludG8gRE5BLg==[Qq]
[c]IFJlc3RyaWN0aW 9uIGVuenltZXM=[Qq]
[f]IEV4Y2VsbGVudC4gUmVzdHJpY3Rpb24gZW56eW1lcyBhcmUgcmVzcG9uc2libGUgZm9yIGN1dHRpbmcgRE5BIGludG8gZnJhZ21lbnRz[Qq]
[c]IEROQSBsaWdhc2U=[Qq]
[f]IE5vLiBETkEgTGlnYXNlIGNyZWF0ZXMgc3VnYXItcGhvc3BoYXRlIGJvbmRzIGJldHdlZW4gYWRqYWNlbnQgZnJhZ21lbnRzIG9mIEROQS4=[Qq]
[q] In the diagram below, a restriction enzyme is represented by
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IENvcnJlY3QhIExldHRlciAmIzgyMjA7ZSYjODIyMTsgaXMgYSByZXN0cmljdGlvbiBlbnp5bWUu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBSZXN0cmljdGlvbiBlbnp5bWVzIGN1dCBvcGVuIEROQS4=[Qq]
[q] In the diagram below, HindIII must be a(n) [hangman] enzyme.
[c]IHJlc3RyaWN0aW9u[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] In the diagram below, the DNA bases with green arrows represent[hangman] ends.
[c]IHN0aWNreQ==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] In the diagram below, a plasmid that’s been treated with a restriction enzyme is shown at
[textentry single_char=”true”]
[c]IG U=[Qq]
[c]WWVzISBMZXR0ZXIgJiM4MjIwO2UmIzgyMjE7IHNob3dzIGEgcGxhc21pZCB0aGF0JiM4MjE3O3MgYmVlbiBjdXQgb3BlbiBieSBhIHJlc3RyaWN0aW9uIGVuenltZS4=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBBIHBsYXNtaWQgdGhhdCA=aGFzbiYjODIxNzt0IGJlZW4gdHJlYXRlZCB3aXRoIGEgcmVzdHJpY3Rpb24gZW56eW1lIGlzIHNob3duIGF0ICYjODIyMDthLiYjODIyMTs=[Qq]
[q] In the diagram below, recombinant DNA first appears at
[textentry single_char=”true”]
[c]IG Y=[Qq]
[f]IFllcyEgJiM4MjIwO0YmIzgyMjE7IGlzIGEgcmVjb21iaW5hbnQgcGxhc21pZC4=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBMb29rIGZvciB0aGUgZmlyc3Qgb2NjdXJyZW5jZSBvZiBodW1hbiBETkEgY29tYmluZWQgd2l0aCBwbGFzbWlkIEROQS4=[Qq]
[q] In the diagram below, the first occurrence of a transgenic organism is at
[textentry single_char=”true”]
[c]IG c=[Qq]
[f]WWVzISBMZXR0ZXIgJiM4MjIwO2cmIzgyMjE7IHNob3dzIGEgcmVjb21iaW5hbnQgb3JnYW5pc20u[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBMb29rIGZvciBhIGNlbGwgdGhhdCBoYXMgYm90aCBiYWN0ZXJpYWwgRE5BIGFuZCBodW1hbiBETkEu[Qq]
[x]
[restart]
[/qwiz]
9. Flashcards: Five Things to Know about Genetic Engineering
[qdeck style=”width: 600px !important; min-height: 450px !important;” bold_text=”false” qrecord_id=”sciencemusicvideosmeister1961-Five things to know about genetic engineering (flashcards, v2.0)”]
[h] Five things to know about genetic engineering
[i]
[start]
[q] Define genetic engineering and recombinant DNA.
[a]
- Genetic engineering is the process of altering the characteristics of an organism by directly manipulating its DNA.
- Recombinant DNA is DNA that is a combination of two (or more) sources. In the context of genetic engineering, the usually means DNA from two species that have been combined into one recombinant DNA molecule.
[q] What are restriction endonucleases, and how did they originate?
[a] Restriction endonucleases are enzymes that can identify a specific sequence within a strand of DNA, and then cut apart that DNA into two pieces. These enzymes evolved in bacteria as a defense against attack by bacteriophages. These enzymes, shown at 4 below, identify viral DNA sequences (the “restriction site” shown at “5” below) and cut viral DNA apart, which keeps the virus from taking over the bacterial cell and converting the cell into a virus factory.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|172809a886910″ question_number=”267″ topic=”6.8.Biotechnology”] It takes several steps to transfer human genes to bacterial cells that can subsequently produce human gene products (such as the human protein insulin). One step involves dealing with introns. Explain what introns are, why they’re a problem, and how they can be removed.
[a] Introns are non-coding sequences of DNA within eukaryotic genes that have to be spliced out before the gene’s RNA can be translated into protein. So, to transfer a human gene to a bacterium to create a gene product, you have to use DNA from which the introns have been removed. You can get this intron-free in two ways. First, you can find cells that produce the desired protein, extract the mRNA that codes for this protein from those cells, and then use reverse transcriptase to create cDNA (complementary DNA) from the mRNA. Alternatively, if you know the amino acid sequence for the protein, you can reverse-engineer DNA that codes for that amino acid sequence.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|171bd06a19910″ question_number=”268″ topic=”6.8.Biotechnology”] Using the diagram below as a stimulus, explain how you can create a recombinant plasmid with a human gene that can be used to produce a human protein. Note: you can assume that introns have already been removed from the human DNA.
[a] To create a recombinant plasmid, extract a plasmid (c) from a bacterial cell. Then cut it open with a restriction enzyme (e) so that its ends are “sticky,” with single-stranded overhangs that can form hydrogen bonds with complementary sequences. Second, use the same restriction enzyme to extract a human gene (f). Third, combine the human DNA fragment and the plasmid DNA. Because they’ve been cut with the same restriction enzyme, they’ll combine by forming hydrogen bonds between their complementary sticky ends (g). Fourth, use DNA ligase (not shown) to create sugar-phosphate bonds between adjacent, unbonded nucleotides. Finally, use transformation to insert the engineered plasmid (g) into a bacterial cell (h). Every time this engineered bacterium reproduces, it will reproduce the plasmid (i). During transcription and translation, the bacterium will synthesize the human protein (j), which can be purified from the bacterial culture and put to use.
[q] Explain how recombinant DNA can be created.
[a] Letter A shows a stretch of DNA, within which is the sequence GAATTC. That sequence is a restriction site (B). It’s recognized by a restriction enzyme (C), which cuts the sugar-phosphate bonds in the DNA between the nucleotide bases guanine and adenine. The result is two DNA fragments (D), each with a “sticky end” (E).
Letter “F” represents a second stretch of DNA that’s been cut with the same restriction enzyme. When that DNA is mixed with the DNA fragments at “D,” the two will combine as is shown in “G.” Adding DNA ligase (i) will create sugar-phosphate bonds between adjacent nucleotides. Once ligase has done its work, you have recombinant DNA (J):
[x] [restart]
[/qdeck]
10. What’s Next?
Proceed to Topic 6.8 Part 2: Gel Electrophoresis, Restriction Fragment Length Analysis, and DNA Fingerprinting (the next tutorial in AP Bio Unit 6)
