1. Introduction: What is a DNA Fingerprint?
|
Fingerprint |
Four DNA Fingerprints |
Unless you’re an identical twin, your DNA is unique. DNA fingerprinting or profiling involves chemically manipulating DNA to create a unique pattern, like the four shown above (one from a crime scene, and one for each of the three suspects in the crime). The pattern can also be a series of numbers, each corresponding to a feature in a person’s DNA.
DNA fingerprinting is widely used in research, and for the past 30 years has been a mainstay of forensics (crime investigation).
2. RFLPs (Restriction Fragment Length Polymorphisms)
An early method for creating a DNA fingerprint was based on restriction fragments. These segments of DNA are created by bacterial enzymes called restriction endonucleases ( restriction enzymes). These enzymes were discussed in the first tutorial in this module (and you should read about them now if you’re doing this tutorial before that one)
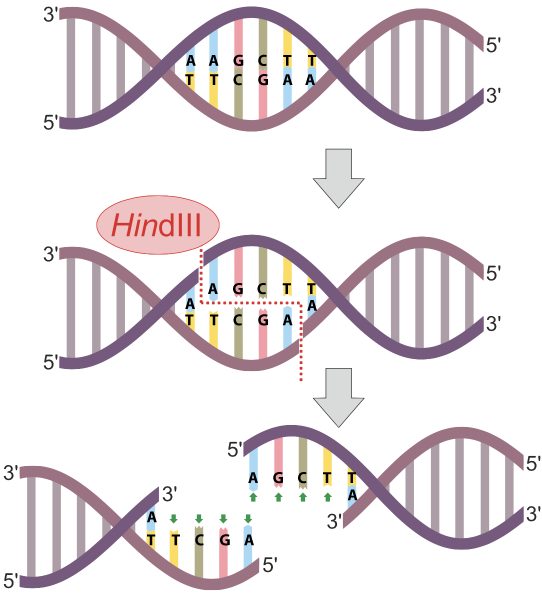
In the diagram at left, the restriction endonuclease HindIII recognizes the nucleotide sequence AAGCTT and cuts the DNA between the two As. Note that because the sequence is a palindrome (it’s the same in the 5′ to 3′ direction on both strands), the cut winds up being double-stranded, and completely severs the DNA.

If you were to take my DNA and put it into a vial with restriction enzymes, my DNA would be cut into fragments. Why? Because along the length of the billions of bases in my DNA, there are going to be a few instances of restriction site sequences. At these restriction sites, the restriction enzyme will cut up my DNA into restriction fragments.
Now, here’s where the creation of restriction fragments ties into DNA profiling and fingerprinting. A restriction site is a sequence of nucleotide bases. While most of the DNA shared by any two humans is the same, there are differences: that’s what makes us genetically distinct. If those differences are within a restriction site, then the number of the restriction fragments created when the DNA from any two people is treated with the same restriction enzyme will also differ, as will the length of those fragments.
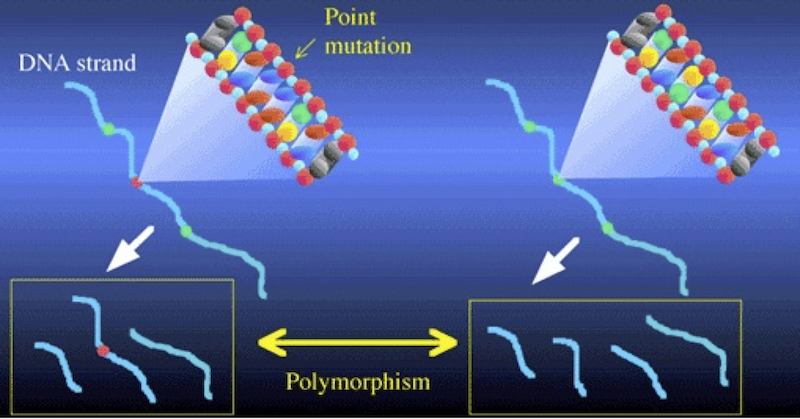
Consider the illustration above. It shows two DNA strands. The sequence on the right has three restriction sites (represented in green), and when cut with the corresponding restriction enzyme, that DNA will be cut into four fragments. In the DNA on the left, there’s been a mutation in the second restriction site (shown in red). As a result, the corresponding restriction enzyme won’t recognize that site, and won’t cut the DNA at that point. Consequently, the left sample of DNA is cut into three fragments, while the right sample is cut into four. Note also how in the DNA strand on the left, the combination of the second and third fragments leads one of the fragments to be considerably longer than any of the others.
These differences between the two DNA strands are polymorphisms. The word polymorphism means “more than one form.” And the basic idea behind RFLP analysis is that differences in restriction sites will result in restriction fragment length polymorphism. The differences are of two kinds: differences in the number of fragments, and differences in the length of the fragments.
3. RFLPS: Checking Understanding
[qwiz random = “false” qrecord_id=”sciencemusicvideosMeister1961-Restriction Enzymes, Fragments, and Polymorphisms (v2.0)”]
[h]Restriction Enzymes, Fragments, and Polymorphisms
[i]
[q] Review: Restriction enzymes were first identified in [hangman], where their function is to fight off [hangman].
[c]IGJhY3Rlcmlh[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IHZpcnVzZXM=[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] HindIII is a(n) [hangman] enzyme.
[c]IHJlc3RyaWN0aW9u[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] The two yellow boxes below both show pieces of [hangman] that have been cut into restriction [hangman].
[c]IEROQQ==[Qq]
[f]IEdyZWF0IQ==[Qq]
[c]IGZyYWdtZW50cw==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] In the diagram below, RFLPs are indicated by the number
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEdyZWF0ISBOdW1iZXIgJiM4MjIwOzImIzgyMjE7IGluZGljYXRlcyBkaWZmZXJlbmNlcyDigJQgcG9seW1vcnBoaXNtcyDigJQgaW4gdGhlIGxlbmd0aHMgb2YgdGhlIHJlc3RyaWN0aW9uIGZyYWdtZW50cyBmcm9tIHR3byBkaWZmZXJlbnQgRE5BIHNhbXBsZXMuIFRoZXNlIGRpZmZlcmVuY2VzIGFyZSBrbm93biBhcyBSRkxQcy4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBSRkxQcyBhcmUgdGhlIGZyYWdtZW50cyB0aGF0IHJlc3VsdCB3aGVuIEROQSBpcyBjdXQgd2l0aCByZXN0cmljdGlvbiBlbnp5bWVzLiBXaGF0JiM4MjE3O3MgdGhlIG9ubHkgdGhpbmcgaW4gdGhpcyBkaWFncmFtIHRoYXQgY291bGQgcmVwcmVzZW50IGZyYWdtZW50cyBvZiBhIGxhcmdlciBwaWVjZSBvZiBETkE/[Qq]
[q] In the diagram below, number 1 represents a(n) [hangman] in a [hangman] site
[c]IG11dGF0aW9u[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IHJlc3RyaWN0aW9u[Qq]
[f]IE5pY2Ugam9iIQ==[Qq]
[x][restart]
[/qwiz]
4. Gel Electrophoresis
In the DNA fingerprinting method described above, creating RFLPs is the first step. Now we want to be able to see those differences. To do that, we take the restriction fragments and subject them to a treatment called gel electrophoresis.
Electrophoresis is “the movement of charged particles in a fluid or gel under the influence of an electric field.” The process involves the type of apparatus shown below. A gel (5), often made of the polysaccharide agarose (a seaweed extract), is placed in an electrophoresis chamber (3). The chamber is essentially a plastic basin that holds a buffer solution that conducts electricity (6). The chamber has positive (4) and negative (2) electrodes, which are attached to a power supply (7).
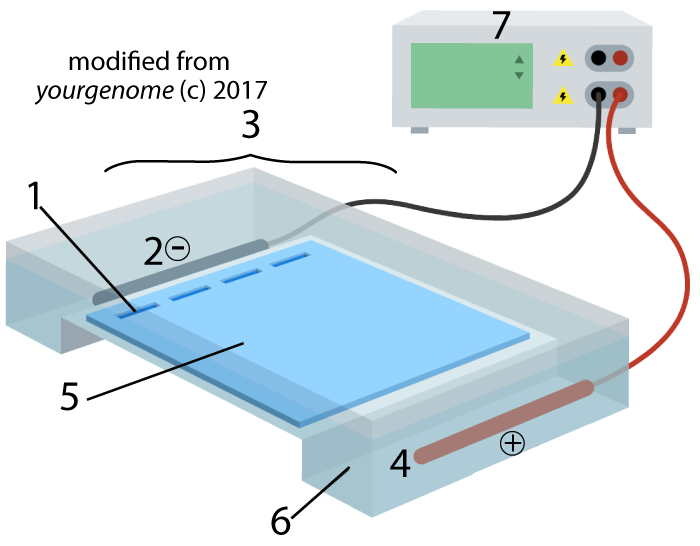
A solution with restriction fragments (also known as a restriction digest) is placed within wells (1), which are indentations in the gel. When you turn the power on, the DNA in the digest will get pushed through the gel, away from the negative electrode (2) and toward the positive electrode (4).
This pushing is related to DNA’s chemistry. DNA’s backbone consists of alternating deoxyribose sugars (the dark blue pentagon with a “D” below) and phosphate groups (the circle with a “P”).
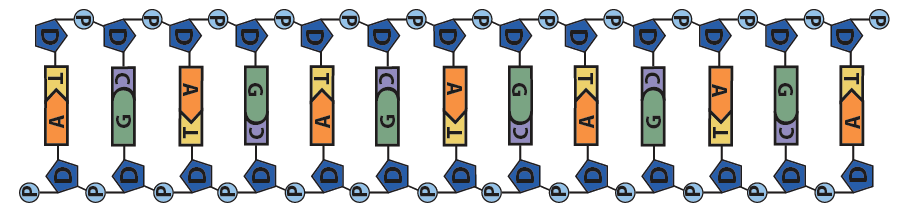
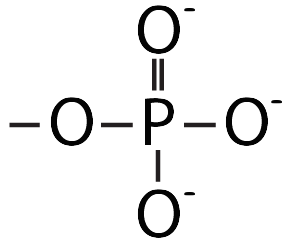
These phosphate groups are negatively charged. As a result, the DNA will be repelled by the negative charge at the negative electrode, and it will be attracted to the positive charge at the positive electrode.
The basic principle of electrophoresis is that small DNA fragments will be impeded by the gel less than large fragments. In other words, large fragments move more slowly than small fragments do, and in a given period, small fragments will move further than large ones.
The image below shows what you would see if you put three different samples of DNA into an electrophoresis chamber. To make sure you’re understanding the explanation above, answer the questions that follow.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Results of electrophoresis (v2.0)”] [h]
Results of electrophoresis
[q] What’s the smallest fragment in the gel shown below?
[textentry single_char=”true”]
[c]ID Y=[Qq]
[f]TmljZS4gJiM4MjIwOzYmIzgyMjE7IHdvdWxkIGhhdmUgdG8gYmUgdGhlIHNtYWxsZXN0IGZyYWdtZW50Lg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBJbiBnZWwgZWxlY3Ryb3Bob3Jlc2lzLCB0aGUgc21hbGxlc3QgZnJhZ21lbnQgaXMgZ29pbmcgdG8gbW92ZSB0aGUgZmFzdGVzdCwgYW5kIGluIGEgZ2l2ZW4gcGVyaW9kLCB0aGUgZmFydGhlc3Qu[Qq]
[q] What’s the largest fragment in the gel shown below?
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]RXhjZWxsZW50OiBOdW1iZXIgMSwgd2hpY2ggbW92ZWQgdGhlIHNob3J0ZXN0IGRpc3RhbmNlLCB3b3VsZCBoYXZlIHRoZSBiZSB0aGUgbGFyZ2VzdCBmcmFnbWVudC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBJbiBnZWwgZWxlY3Ryb3Bob3Jlc2lzLCB0aGUgbGFyZ2VzdCBmcmFnbWVudCBpcyBnb2luZyB0byBtb3ZlIHRoZSBzbG93ZXN0LCBhbmQgaW4gYSBnaXZlbiBwZXJpb2QsIHRoZSBzbWFsbGVzdCBkaXN0YW5jZS4=[Qq]
[q] Assuming that all three samples below consist of linear DNA (not a circular plasmid) and were exposed to the same restriction enzyme, which sample had one restriction site?
[textentry single_char=”true”]
[c]IE I=[Qq]
[f]TmljZTogU2FtcGxlIEIgd2FzIGN1dCBpbnRvIHR3byBmcmFnbWVudHMuIFRoYXQgbWVhbnMgdGhhdCB0aGUgRE5BIGluIHNhbXBsZSBCIGhhZCBvbmUgcmVzdHJpY3Rpb24gc2l0ZSwgYW5kIGEgcmVzdHJpY3Rpb24gZW56eW1lIG1hZGUgYSBjdXQgYXQgdGhhdCBzaXRlLCBkaXZpZGluZyB0aGUgRE5BIGludG8gdHdvIGZyYWdtZW50cy4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJZiB0aGUgRE5BIGhhcyBvbmUgcmVzdHJpY3Rpb24gc2l0ZSwgaXQmIzgyMTc7bGwgYmUgY3V0IGludG8gdHdvIGZyYWdtZW50cy4=[Qq]
[q] Assuming that all three samples below consist of linear DNA (not a circular plasmid) which sample had two restriction sites?
[textentry single_char=”true”]
[c]IE M=[Qq]
[f]R3JlYXQgam9iOiBTYW1wbGUgQyB3YXMgY3V0IGludG8gdGhyZWUgZnJhZ21lbnRzLiBUaGF0IG1lYW5zIHRoYXQgdGhlIEROQSBpbiBzYW1wbGUgQyBoYWQgdHdvIHJlc3RyaWN0aW9uIHNpdGVzLiBUaGUgcmVzdHJpY3Rpb24gZW56eW1lIG1hZGUgYSBjdXQgYXQgZWFjaCBvZiB0aG9zZSBzaXRlcywgZGl2aWRpbmcgdGhlIEROQSBpbnRvIHRocmVlIGZyYWdtZW50cy4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJZiB0aGUgRE5BIGhhcyBvbmUgcmVzdHJpY3Rpb24gc2l0ZSwgaXQmIzgyMTc7bGwgYmUgY3V0IGludG8gdHdvIGZyYWdtZW50cy4gSWYgaXQgaGFzIHR3byByZXN0cmljdGlvbiBzaXRlcywgaXQmIzgyMTc7bGwgYmUgY3V0IGludG8gdGhyZWUgZnJhZ21lbnRzLg==[Qq]
[q] Assuming that all three samples below consist of linear DNA (not a circular plasmid) which sample had no restriction sites for that restriction enzyme?
[textentry single_char=”true”]
[c]IE E=[Qq]
[f]VGVycmlmaWMuIFNhbXBsZSBBIGlzIG9uZSBmcmFnbWVudCwgcmVwcmVzZW50ZWQgYnkgdGhlIGxpbmUgYXQgMS4gVGhhdCBtZWFucyB0aGF0IHRoZSBETkEgaW4gc2FtcGxlIEEgaGFkIG5vIHJlc3RyaWN0aW9uIHNpdGVzLCBhbmQgd2FzbiYjODIxNzt0IGN1dCBhcGFydC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJZiB0aGUgRE5BIGhhcyBhbnkgcmVzdHJpY3Rpb24gc2l0ZXMsIGl0JiM4MjE3O2xsIGJlIGN1dCBpbnRvIHNtYWxsZXIgZnJhZ21lbnRzLiBJZiBpdCByZW1haW5zIG9uZSBwaWVjZSB0aGF0IG1lYW5zIHRoYXQgbm90aGluZyBjdXQgaXQgYXBhcnQu[Qq]
[/qwiz]
5. RFLPs and Plasmid Analysis
What’s shown above gives you the basic idea behind RFLP analysis. In a typical AP Biology lab experience, you might take a plasmid or viral DNA, digest it with restriction enzymes, and run the resulting fragments through gel electrophoresis.
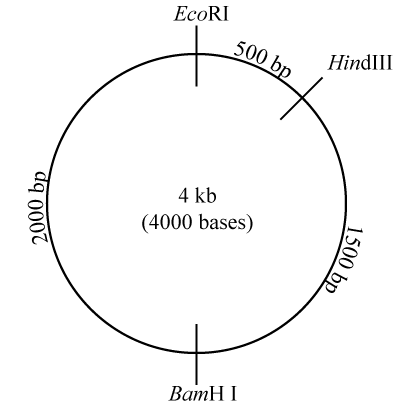 Take a look at the diagram to your right, which represents a plasmid that’s 4000 base pairs long (“4 kb” means four kilobases or 4000 bases). On this plasmid are three restriction sites: one for the restriction enzyme EcoRI, one for HindIII, and one for BamH I. In case you were curious about this, the names of these restriction enzymes include information about the bacteria where the enzyme was first isolated, the specific bacterial strain, and the order of its discovery. EcoRI, for example, was the first restriction enzyme identified in the R strain of E. coli.
Take a look at the diagram to your right, which represents a plasmid that’s 4000 base pairs long (“4 kb” means four kilobases or 4000 bases). On this plasmid are three restriction sites: one for the restriction enzyme EcoRI, one for HindIII, and one for BamH I. In case you were curious about this, the names of these restriction enzymes include information about the bacteria where the enzyme was first isolated, the specific bacterial strain, and the order of its discovery. EcoRI, for example, was the first restriction enzyme identified in the R strain of E. coli.
What this image is telling you is that the restriction site for HindIII is 500 base pairs (bp) away from the restriction site for EcoRI. If you continue moving clockwise, in another 1500 bp you’ll encounter the restriction site for BamH I. Move clockwise another 2000 bp, and you’ll be back at the restriction site for EcoRI.
Note that because the plasmid is a closed loop, a cut at one restriction site merely opens the loop, and won’t create multiple fragments. For example, if the plasmid shown on the right were cut with restriction enzyme EcoR1, the result would be a single piece of DNA that would be 4kb (4000 bases) long.
Work through the questions that follow to get a sense of how to think about this material.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Restriction Sites and Restriction Fragments in Plasmid DNA (v2.0)”]
[h]Restriction Sites and Restriction Fragments in Plasmid DNA
[i]
[q] If the plasmid below is treated with EcoRI and HindIII, the result will be[hangman] restriction fragment(s). The shorter one will be [hangman] bp long, and the longer one will be [hangman] bp long.
[c]IDI=[Qq]
[f]IEdvb2Qh[Qq]
[c]IDUwMA==[Qq]
[f]IEdyZWF0IQ==[Qq]
[c]IDM1MDA=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] If the plasmid below is treated with BamH I and HindIII, the result will be [hangman] restriction fragment(s). The shorter one will be [hangman] bp long, and the longer one will be [hangman] bp long.
[c]IDI=[Qq]
[f]IEdyZWF0IQ==[Qq]
[c]IDE1MDA=[Qq]
[f]IENvcnJlY3Qh[Qq]
[c]IDI1MDA=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q multiple_choice = “true] The image to the left, below, shows a 20 kb plasmid with several restriction sites. The image on the right shows the results of electrophoresis following various combinations of restriction enzymes. Which lane shows the gel that would result if the plasmid were digested with BamH I?
[c]IEHCoCDCoCDCoCA=[Qq][c]IELCoCDC oCDCoMKg[Qq][c]IEM=
Cg==[Qq][f]IE5vLiBUaGUgc2luZ2xlIGxpbmUgaW4gbGFuZSAmIzgyMjA7QSYjODIyMTsgaW5kaWNhdGVzIHRoYXQgb25seSBvbmUgZnJhZ21lbnQgd291bGQgcmVzdWx0IGFuZCB0aGF0IHRoaXMgZnJhZ21lbnQgd291bGQgYmUgMjAga2IgaW4gc2l6ZS4gSWYgeW91IGN1dCB0aGlzIHBsYXNtaWQgd2l0aCBCYW1IMSwgeW91JiM4MjE3O2xsIG1ha2UgMyBjdXRzLiBOb3csIGp1c3QgZmlndXJlIG91dCBob3cgYmlnIGVhY2ggb2YgdGhlIGZyYWdtZW50cyB3b3VsZCBiZSwgYW5kIHlvdSYjODIxNztsbCBoYXZlIHlvdXIgYW5zd2VyLg==[Qq]
[f]IEV4Y2VsbGVudC4gQ3V0dGluZyB0aGlzIHBsYXNtaWQgd2l0aCBCYW1ISSB3aWxsIHJlc3VsdCBpbiB0aHJlZSBmcmFnbWVudHMsIGFuZCB0aGV5JiM4MjE3O2xsIGJlIDExIGtiLCA2IGtiLCBhbmQgMyBLYiBpbiBzaXplLCB3aGljaCBpcyBqdXN0IHdoYXQgaXMgc2hvd24gaW4gdGhpcyBnZWwu[Qq]
[f]IE5vLiBUaGVyZSBhcmUgdGhyZWUgbGluZXMgaW4gbGFuZSBDLCBhbmQgb25lIG9mIHRob3NlIGNvbnNpc3RzIG9mIHR3byBmcmFnbWVudHMuIFRoZSBmaXJzdCBsaW5lIGNvbnNpc3RzIG9mIGEgcmVzdHJpY3Rpb24gZnJhZ21lbnQgdGhhdCYjODIxNztzIDgga2IgbG9uZzsgdGhlIHNlY29uZCBoYXMgYSBmcmFnbWVudCB0aGF0JiM4MjE3O3MgNiBrYiBsb25nLCBhbmQgdGhlIHRoaXJkIGNvbnNpc3RzIG9mIHR3byBmcmFnbWVudHMsIGJvdGggb2Ygd2hpY2ggYXJlIDMga2IuIFdoaWNoIGVuenltZXMgd291bGQgcHJvZHVjZSB0aGF0IHBhdHRlcm4gb2YgZnJhZ21lbnRzPw==[Qq]
[q multiple_choice = “true] The image to the left, below, shows a 20 kb plasmid with several restriction sites. The image on the right shows the results of electrophoresis following various combinations of restriction enzymes. Which lane shows the gel that would result if the plasmid were digested with EcoRI?
[c]IEHCoCDC oCDCoA==[Qq][c]IELCoCDCoCDCoA==[Qq][c]IEM=
Cg==[Qq][f]IE5pY2UgSm9iISBUaGUgc2luZ2xlIGxpbmUgaW4gbGFuZSAmIzgyMjA7QSYjODIyMTsgaW5kaWNhdGVzIHRoYXQgb25seSBvbmUgZnJhZ21lbnQgd291bGQgcmVzdWx0IGFuZCB0aGF0IHRoaXMgZnJhZ21lbnQgd291bGQgYmUgMjAga2IgaW4gc2l6ZS4=[Qq]
[f]IE5vLiBUaGVyZSYjODIxNztzIG9ubHkgb25lIHJlc3RyaWN0aW9uIHNpdGUgcmVjb2duaXplZCBieQ==IEVjbw==IFJJLiBJZiB0aGUgcGxhc21pZCBpcyBjdXQgb3ZlciBhdCB0aGF0IHBvaW50LCBob3cgbWFueSBmcmFnbWVudHMgd291bGQgcmVzdWx0Pw==[Qq]
[f]IE5vLiBUaGVyZSBhcmUgdGhyZWUgbGluZXMgaW4gbGFuZSBDLCBhbmQgb25lIG9mIHRob3NlIGNvbnNpc3RzIG9mIHR3byBmcmFnbWVudHMuIEJ1dCB0aGVyZSYjODIxNztzIG9ubHkgb25lIHJlc3RyaWN0aW9uIHNpdGUgcmVjb2duaXplZCBieQ==IEVjbw==IFJJLiBJZiB0aGUgcGxhc21pZCBpcyBjdXQgb3ZlciBhdCB0aGF0IHBvaW50LCBob3cgbWFueSBmcmFnbWVudHMgd291bGQgcmVzdWx0Pw==[Qq]
[q multiple_choice = “true] The image to the left, below, shows a 20 kb plasmid with several restriction sites. The image on the right shows the results of electrophoresis following various combinations of restriction enzymes. Which lane shows the gel that would result if the plasmid were digested with Bam HI and EcoRI?
[c]IEHCoCDCoCDCoA==[Qq][c]IELCoCDCoCDCoA==[Qq][c]IE M=
Cg==[Qq][f]IE5vLiBUaGUgc2luZ2xlIGxpbmUgaW4gbGFuZSAmIzgyMjA7QSYjODIyMTsgaW5kaWNhdGVzIHRoYXQgdGhlcmUmIzgyMTc7cyBvbmx5IG9uZSBmcmFnbWVudCBhbmQgdGhhdCB0aGlzIGZyYWdtZW50IHdvdWxkIGJlIDIwIGtiIGluIHNpemUuIEhvdyBjb3VsZCB0aGF0IG9jY3VyIGlmIGJvdGggQmFtIEhJIA==YW5kIA==[Qq]Eco R I were used to digest the plasmid?
[f]IE5vLiBMYW5lIEIgaW5kaWNhdGVzIG9uZSBmcmFnbWVudCBvZiAxMSBrYiwgYSBzZWNvbmQgb2YgNiBrYiwgYW5kIGEgM3JkIG9mIDMga2Iu[Qq]
[f]IEV4Y2VsbGVudC4gVGhlcmUgYXJlIHRocmVlIGxpbmVzIGluIGxhbmUgQywgYW5kIG9uZSBvZiB0aG9zZSBjb25zaXN0cyBvZiB0d28gZnJhZ21lbnRzLiBUaGF0JiM4MjE3O3MgZXhhY3RseSB3aGF0IHlvdSYjODIxNztkIGV4cGVjdCBpZiBib3RowqA=QmFtIEhJIA==YW5kIA==[Qq]Eco R I were used to digest the plasmid.
[/qwiz]
What’s Next?
The DNA fingerprints used in modern forensics are created using much more complex and accurate techniques than what we described above (which only works with short pieces of DNA, such as that in plasmids and viruses). To learn about these techniques (which are beyond the scope of a typical AP Bio curriculum), please consult this tutorial in our College Biology curriculum.
Proceed to Topic 6.8, Part 3: Amplifying DNA by PCR (the Polymerase Chain Reaction), the next tutorial in AP Bio Unit 6.


