1. Introduction
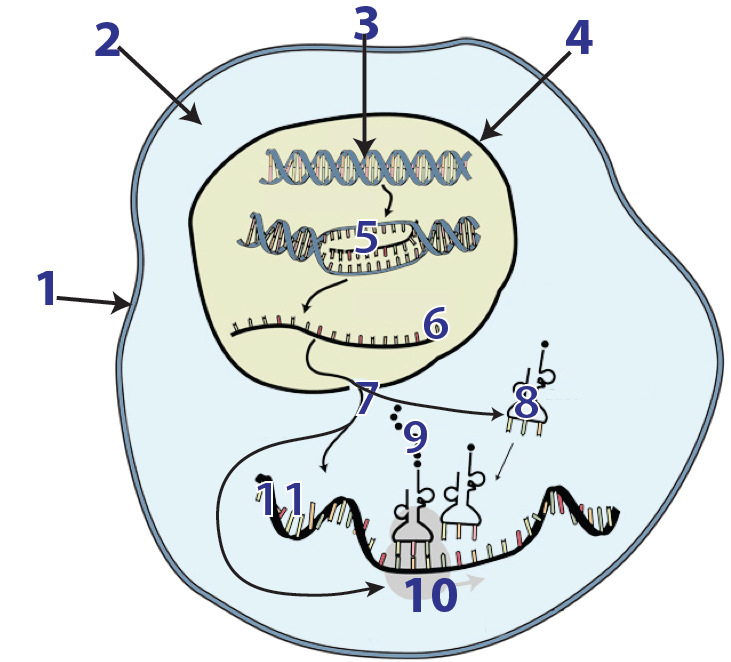
In previous tutorials in Unit 6, we learned about transcription and the genetic code. In this tutorial, we’ll look at the cellular mechanisms involved in translation. During translation, which is also known as protein synthesis, ribosomes (10) “read” an mRNA message (6 and 11). With the assistance of tRNAs (8), the ribosome translates the mRNA message into a polypeptide (9): a sequence of amino acids that makes up the primary structure of a protein.
Transcription and translation link genotype with phenotype. Organisms inherit genes. The expression of those genes leads to an organism’s phenotype. In other words, the code for each protein can be traced back to an organism’s DNA, which gets transcribed into RNA, which gets translated into protein. In what follows, we’ll focus on the last information transfer (RNA to protein). Let’s go.
2. Protein Structure and Function Review
In Unit 1 of this course, you learned about the structure and function of proteins. Here’s a quick review. If you feel that you remember that material well, jump down to the protein review quiz that follows this reading.
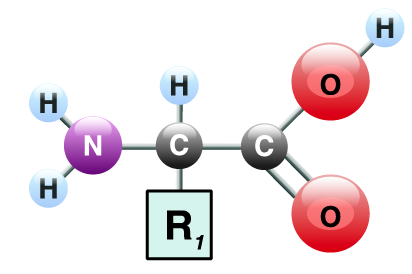
1. Proteins are polymers of amino acids. As shown below, each amino acid consists of a central carbon atom. The central carbon is bonded to an amino group (—NH2) on one side, a carboxylic acid group (—COOH) on the other side, and a side chain (also known as an “R-group”). The side chains vary in their composition and chemistry and can be polar/hydrophilic, non-polar/hydrophobic, acidic, or basic.
2. There are 20 amino acids. You can think of them as an alphabet. In the same way that English uses the 26 letters of the alphabet to create hundreds of thousands of words, cells use various combinations of amino acids to create hundreds of thousands of proteins. As a result, proteins are the most diverse macromolecule.
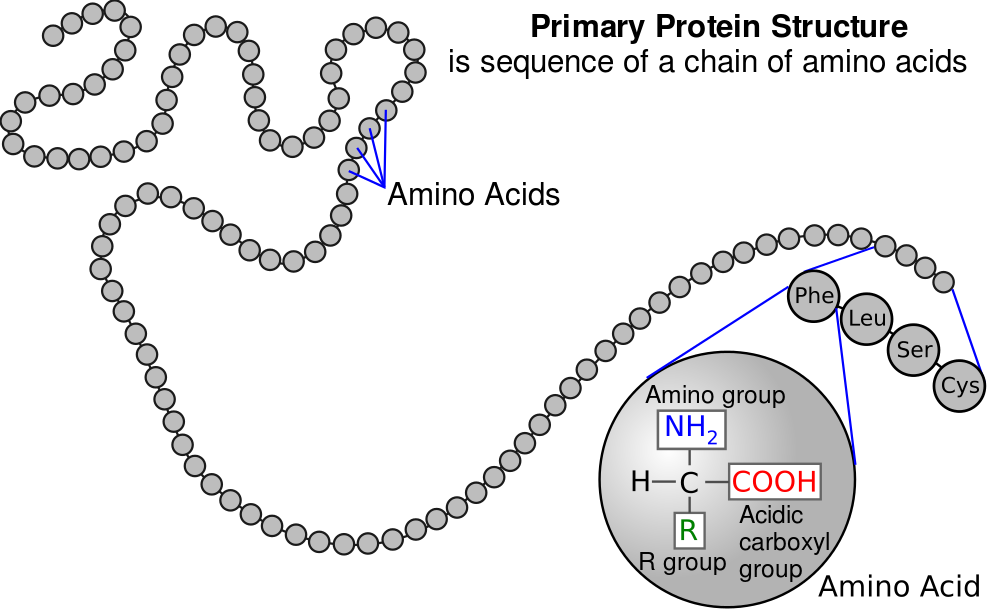
3. In diagrams about proteins and protein synthesis, amino acids are represented by three-letter abbreviations. In what’s above, Phe represents the amino acid phenylalanine, leu represents leucine, and so on.
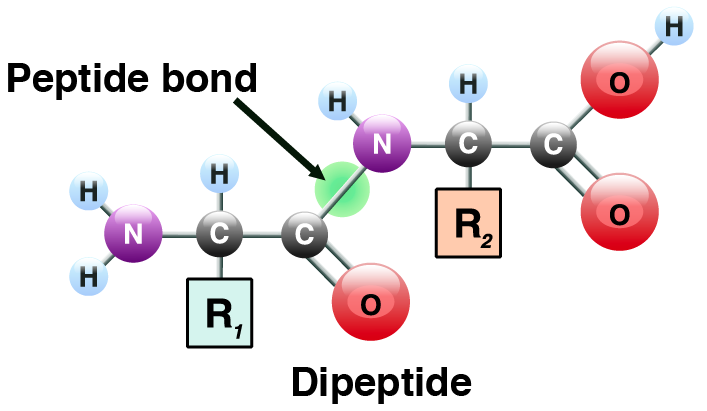
4. Amino acids in proteins are joined by peptide bonds (find the bond indicated by the green circle above). Two linked amino acids form a dipeptide (see above). A chain of amino acids that are linked together is known as a polypeptide (see # 1 in the diagram below). A polypeptide can as short as a few amino acids, or hundreds of amino acids long.
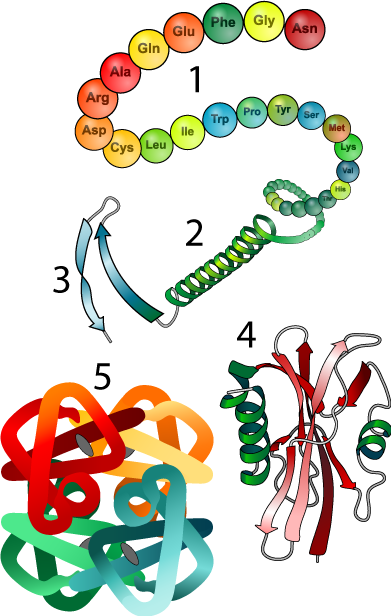 5. Interactions between amino acids cause a polypeptide to fold into a specific shape. This leads to the notion of levels of protein structure. For a full review of protein structure, you can watch this video, and or go back to this tutorial in unit 1.
5. Interactions between amino acids cause a polypeptide to fold into a specific shape. This leads to the notion of levels of protein structure. For a full review of protein structure, you can watch this video, and or go back to this tutorial in unit 1.
- Primary structure is the genetically determined, linear sequence of amino acids in a polypeptide chain (number 1 in the diagram to your right).
- Secondary structure (2) results from hydrogen bonds that form between carbonyl and hydroxyl groups within the polypeptide backbone. This results in alpha helices (think of a corkscrew), or beta-pleated sheets (think of a piece of paper with many parallel folds).
- Tertiary structure results from interactions between amino acid side chains, resulting in complex turns and loops. One such interaction is responsible for the hairpin turn at 3. Many proteins, such as the one shown at 4 are tertiary
- Quaternary (4th-level) structure results from interactions between separate polypeptide chains.
6. The shape that emerges from these interactions determines a protein’s function. For example, the enzyme shown below will only be able to interact with its substrate if its active site (the indentation on the upper right) has a specific shape.

3. Protein Chemistry Review Quiz
[qwiz random = “false” qrecord_id=”sciencemusicvideosMeister1961-Protein Chemistry Review (v2.0)”][h]
Review Quiz: Protein Chemistry and Structure
[i]
[q] The monomers of proteins are [hangman] [hangman].
[c]YW1pbm8=[Qq]
[c]YWNpZHM=[Qq]
[q json=”true” xx=”2″ json=”true” multiple_choice=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f7971251f1241″ question_number=”3″] In the diagram below, which number indicates the amino group?
[c]IDEg[Qq][c]ID Ig[Qq][c]IDMg[Qq][c]IDQ=[Qq][f]IEV4Y2VsbGVudCEgJiM4MjIwOzEmIzgyMjE7IGlzIHRoZSBjZW50cmFsIGNhcmJvbg==
Cg==[Qq][f]IEdyZWF0IGpvYi4gJiM4MjIwOzImIzgyMjE7IGlzIGFuIGFtaW5vIGdyb3VwLg==[Qq]
[f]IE5vLiAmIzgyMjA7MyYjODIyMTsgaXMgdGhlIGNhcmJveHlsaWMgYWNpZA==[Qq]
[f]IE5vLiAmIzgyMjA7MSYjODIyMTsgaXMgdGhlIGNlbnRyYWwgY2FyYm9u[Qq]
[q json=”true” xx=”2″ json=”true” multiple_choice=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f789af0d92241″ question_number=”5″] In the diagram below, which number indicates the part that varies from one amino acid to the next?
[c]IDEg[Qq][c]IDIg[Qq][c]IDMg[Qq][c]ID Q=
Cg==[Qq][f]IEV4Y2VsbGVudCEgJiM4MjIwOzEmIzgyMjE7IGlzIHRoZSBjZW50cmFsIGNhcmJvbg==[Qq]
[f]IE5vLiAmIzgyMjA7MiYjODIyMTsgaXMgYW4gYW1pbm8gZ3JvdXAu[Qq]
[f]IE5vICYjODIyMDszJiM4MjIxOyBpcyB0aGUgY2FyYm94eWxpYyBhY2lk[Qq]
[f]IFllcyAmIzgyMjA7NCYjODIyMTsgaXMgdGhlICYjODIyMDtSIGdyb3VwLCYjODIyMTsgdGhlIHBhcnQgdGhhdCBjYW4gdmFyeSBmcm9tIG9uZSBhbWlubyBhY2lkIHRvIHRoZSBuZXh0Lg==[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f7762f29fca41″ question_number=”8″] In the diagram below, which letter indicates a peptide bond?
[textentry single_char=”true”]
[c]IG M=[Qq]
[f]IEV4Y2VsbGVudCEgTGV0dGVyICYjODIyMDtjJiM4MjIxOyBpcyBwb2ludGluZyB0byBhIHBlcHRpZGUgYm9uZA==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBQZXB0aWRlIGJvbmRzIGNvbm5lY3Qgb25lIGFtaW5vIGFjaWQgdG8gdGhlIG5leHQuIFRyeSB0byBmaW5kIHdoYXQmIzgyMTc7cyBsZWZ0IG9mIGEgY2FyYm94eWwgZ3JvdXAgaW4gb25lIGFtaW5vIGFjaWQgYW5kIHdoYXQmIzgyMTc7cyBsZWZ0IG9mIGFuIGFtaW5vIGFjaWQgaW4gYW5vdGhlciBhbWlubyBhY2lkLiBUaGUgY29ubmVjdGlvbiBiZXR3ZWVuIHRoZSB0d28gaXMgYSBwZXB0aWRlIGJvbmQu[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f76fc80949a41″ question_number=”9″] In the diagram below, which letter indicates a polar R group?
[textentry single_char=”true”]
[c]IG Y=[Qq]
[f]IEF3ZXNvbWUhIExldHRlciAmIzgyMjA7ZiYjODIyMTsgaXMgYSBwb2xhciBSIGdyb3VwLiBZb3UgY2FuIHRlbGwgYmVjYXVzZSBpdCBoYXMgYSBoeWRyb3h5bCBncm91cCBhdCBpdHMgZW5kLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBQb2xhciBSIGdyb3VwcyBoYXZlIHBvbGFyIGZ1bmN0aW9uYWwgZ3JvdXBzLiBMb29rIGZvciB0aGUgUiBncm91cCB0aGF0IGhhcyBhIGh5ZHJveHlsIGdyb3VwICjigJRPSCku[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f7631f08a1e41″ question_number=”11″] In the diagram below, which letter indicates a basic R group?
[textentry single_char=”true”]
[c]IG g=[Qq]
[f]IEF3ZXNvbWUhIExldHRlciAmIzgyMjA7aCYjODIyMTsgaXMgYSBiYXNpYyBSIGdyb3VwLiBZb3UgY2FuIHRlbGwgYmVjYXVzZSBpdCBoYXMgYW4gYW1pbm8gZnVuY3Rpb25hbCBncm91cC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBCYXNpYyBSIGdyb3VwcyBoYXZlIGFuIGFtaW5vIGZ1bmN0aW9uYWwgZ3JvdXAgYXR0YWNoZWQgdG8gdGhlbS4gTG9vayBmb3IgdGhlIFIgZ3JvdXAgdGhhdCBoYXMgYW4gYW1pbm8gZnVuY3Rpb25hbCBncm91cCAo4oCUTkg=Mw==KS4=[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f741371b79a41″ question_number=”16″] The diagram below shows a secondary structure known as a(n) [hangman] [hangman].
[c]YWxwaGE=[Qq]
[c]aGVsaXg=[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f73acffac6a41″ question_number=”17″] the diagram below shows a type of secondary structure called a beta [hangman] [hangman].
[c]cGxlYXRlZA==[Qq]
[c]c2hlZXQ=[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f73468da13a41″ question_number=”18″] In the diagram below, the polypeptide backbone is indicated by number
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IFllcy4gVGhlIHBvbHlwZXB0aWRlIGJhY2tib25lIGlzIGluZGljYXRlZCBieSBudW1iZXIgJiM4MjIwOzEuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIG51bWJlciB0aGF0JiM4MjE3O3Mgbm90IHBvaW50aW5nIHRvIGFueSBvZiB0aGUgaW50ZXJhY3Rpbmcgc2lkZSBjaGFpbnMu[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f72e01b960a41″ question_number=”19″] In the diagram below, a hydrogen bond between two side chains is indicated by number
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IE5pY2UuIE51bWJlciAmIzgyMjA7MiYjODIyMTsgc2hvd3MgdHdvIHNpZGUgY2hhaW5zIHRoYXQgYXJlIGh5ZHJvZ2VuIGJvbmRpbmcgd2l0aCBvbmUgYW5vdGhlciwgY2F1c2luZyBhIGJlbmQgaW4gdGhlIHBvbHlwZXB0aWRlIGJhY2tib25lLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIG51bWJlciB0aGF0JiM4MjE3O3Mgbm90IHBvaW50aW5nIHRvIGFueSBvZiB0aGUgaW50ZXJhY3Rpbmcgc2lkZSBjaGFpbnMu[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f7210e373c641″ question_number=”21″] In the diagram below, hydrophobic clustering is indicated by number
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IFdheSB0byBnbyEgTnVtYmVyICYjODIyMDs0JiM4MjIxOyBzaG93cyBoeWRyb3Bob2JpYyBjbHVzdGVyaW5nLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBIeWRyb3Bob2JpYyBjbHVzdGVyaW5nIGludm9sdmVzIG5vbi1wb2xhciwgaHlkcm9waG9iaWMgc2lkZS1jaGFpbnMuIExvb2sgZm9yIHR3byBzaWRlIGNoYWlucyB0aGF0IGhhdmUgbm9uLXBvbGFyIGZ1bmN0aW9uYWwgZ3JvdXBzLCBhbmQgYXJlIGNsdXN0ZXJpbmcgdG9nZXRoZXIu[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f713d0339ba41″ question_number=”23″] In the diagram below, primary structure is indicated by number
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IEV4Y2VsbGVudC4gUHJpbWFyeSBzdHJ1Y3R1cmUgaXMgaW5kaWNhdGVkIGJ5IG51bWJlciAmIzgyMjA7MS4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBQcmltYXJ5IHN0cnVjdHVyZSBpcyB0aGUgc2VxdWVuY2Ugb2YgYW1pbm8gYWNpZHMuIFdoaWNoIG51bWJlciBzaG93cyB0aGF0IHNlcXVlbmNlICh3aXRob3V0IGFueSBvZiB0aGUgZm9sZGluZyBhbmQgYmVuZGluZyB0aGF0IGVtZXJnZXMgYXQgaGlnaGVyIGxldmVscyBvZiBzdHJ1Y3R1cmU/[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f70d1e916c241″ question_number=”24″] In the diagram below, an alpha helix is indicated by number
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IFRlcnJpZmljLiBBbiBhbHBoYSBoZWxpeCAoYSBzZWNvbmRhcnkgbGV2ZWwgb2YgcHJvdGVpbiBzdHJ1Y3R1cmUpIGlzIHNob3duIGF0IG51bWJlciAmIzgyMjA7Mi4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBTZWNvbmRhcnkgc3RydWN0dXJlIGludm9sdmVzIGh5ZHJvZ2VuIGJvbmRpbmcgYmV0d2VlbiB0aGUgYW1pbm8gYW5kIGNhcmJvbnlsIGZ1bmN0aW9uYWwgZ3JvdXBzIHdpdGhpbiBhIHByb3RlaW4mIzgyMTc7cyBwb2x5cGVwdGlkZSBiYWNrYm9uZS4gVHdvIHByaW1hcnkgc2Vjb25kYXJ5IGxldmVsIHN0cnVjdHVyZXMgYXJlIGFscGhhIGhlbGljZXMgYW5kIGJldGEgcGxlYXRlZCBzaGVldHMuIFRoZSBoZWxpeCBpcyBsaWtlIGEgY29ya3NjcmV3ICh3aGljaCBzaG91bGQgaGVscCB5b3UgZmluZCBpdCku[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f703a9211de41″ question_number=”25″] In the diagram below, a protein with a tertiary structure (but not quaternary structure) is indicated by number
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IE5pY2UuIEEgcHJvdGVpbiB3aXRoIGEgdGVydGlhcnkgc3RydWN0dXJlIGlzIGluZGljYXRlZCBieSBudW1iZXIgJiM4MjIwOzQuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUZXJ0aWFyeSBzdHJ1Y3R1cmUgaXMgd2hhdCBhcmlzZXMgYnkgYm9uZGluZyBiZXR3ZWVuIHNpZGUgY2hhaW5zIHdpdGhpbiBhIHNpbmdsZSBwb2x5cGVwdGlkZSBjaGFpbiwgd2hpY2ggcmVzdWx0cyBpbiBhIHZlcnkgY29tcGxleCB0aHJlZS1kaW1lbnNpb25hbCBzaGFwZS4gSG93ZXZlciwgdGhlIHJlc3VsdGluZyBwcm90ZWluIGNvbnNpc3RzIG9mIGp1c3Qgb25lIHBvbHlwZXB0aWRlIGNoYWluLCBub3QgbXVsdGlwbGUgY2hhaW5zLiBXaGljaCBwYXJ0IG9mIHRoZSBkaWFncmFtIGZpdHMgdGhhdCBkZXNjcmlwdGlvbj8=[Qq]
[q json=”true” xx=”2″ json=”true” dataset_id=”SMV_Protein Synthesis_protein chemistry review|f6f82a2668241″ question_number=”26″] In the diagram below, a protein with a quaternary structure is indicated by number
[textentry single_char=”true”]
[c]ID U=[Qq]
[f]IFBlcmZlY3QuIEEgcHJvdGVpbiB3aXRoIGEgcXVhdGVybmFyeSBzdHJ1Y3R1cmUgaXMgaW5kaWNhdGVkIGJ5IG51bWJlciAmIzgyMjA7NS4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBRdWF0ZXJuYXJ5IHN0cnVjdHVyZSBpcyB3aGF0IGFyaXNlcyBieSBpbnRlcmFjdGlvbnMgYmV0d2VlbiBtdWx0aXBsZSBwb2x5cGVwdGlkZSBjaGFpbnMuIEJvbmRzIGJldHdlZW4gdGhlc2UgY2hhaW5zIGxlYWQgdGhlbSB0byBmb3JtIGEgbXVsdGktcG9seXBlcHRpZGUgY2hhaW4gcHJvdGVpbi4gV2hpY2ggbnVtYmVyIHNob3dzIG11bHRpcGxlIHBvbHlwZXB0aWRlIGNoYWlucz8=[Qq]
[x][restart]
[/qwiz]
4.mRNA, tRNA, and Ribosomes
Ribosomes can be thought of as nano-robots that can read mRNA to make any protein that the cell can specify. How does the ribosome perform this feat? The process involves interaction and information flow between three types of RNA: mRNA, tRNA, and rRNA.
4a. mRNA
mRNA is a polymer of RNA nucleotides. As explained in the previous tutorial about the genetic code, mRNA’s function is to specify the order of amino acids in a polypeptide.
Within the coding portion of mRNA, every three nucleotides make up a codon. Each codon codes for one amino acid.
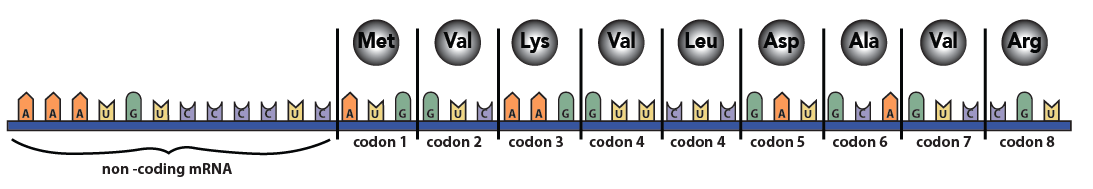
Some codons serve as punctuation. The codon “AUG,” for example, signals the start of a polypeptide chain. Three other codons indicate the chain’s end.
4b. Transfer RNAs (tRNAs) bring amino acids to the ribosome.
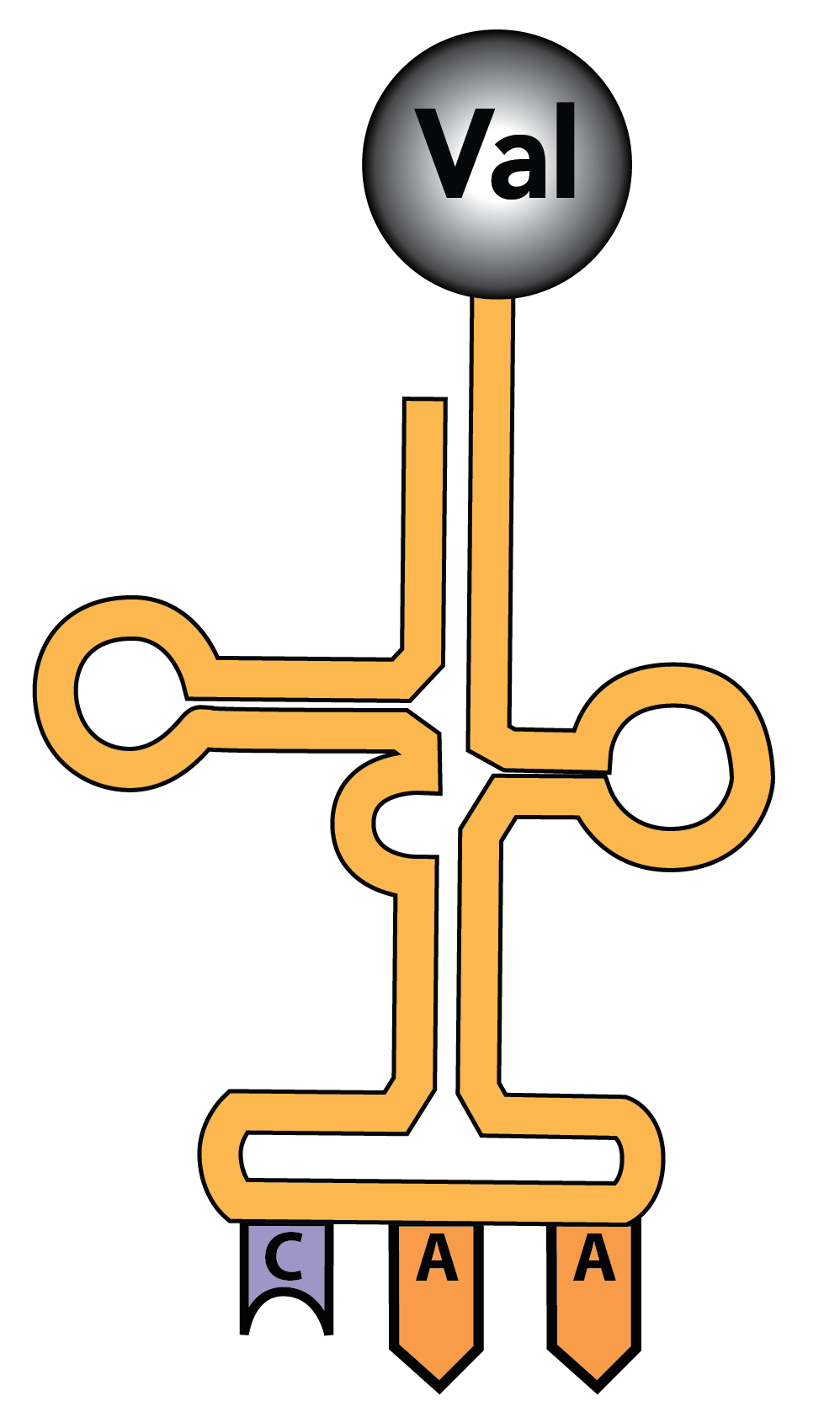
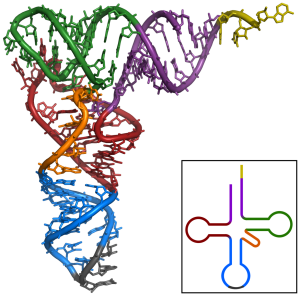
tRNAs are also RNA polymers. They’re generally between 75 and 90 RNA nucleotides long. But unlike mRNAs, which are linear, hydrogen bonding between nucleotides within a tRNA causes it to fold up. The resulting shape is often represented as a type of cloverleaf, as shown on the left. A more realistic model showing some of the base pairings between complementary nucleotides is shown on the right.
On the bottom of the tRNA is an anti-codon: 3 RNA nucleotides that complement the codons in RNA. In the diagram to the left, the anti-codon is “CAA.” This would bind with the codon GUU on an mRNA.
The top of the tRNA has an amino acid binding site. In the tRNA shown on the left, the amino acid is “Val,” short for valine.
The diagram below shows tRNAs (5) in the context of protein synthesis. “1” is the ribosome. Note how the anticodon GGG (at “3”) is binding with the codon CCC on mRNA (2). That same tRNA is carrying the amino acid gly (glycine), which the ribosome will soon attach to the growing polypeptide chain (7) with a peptide bond (6). We’ll explain more of the details of how this happens below.
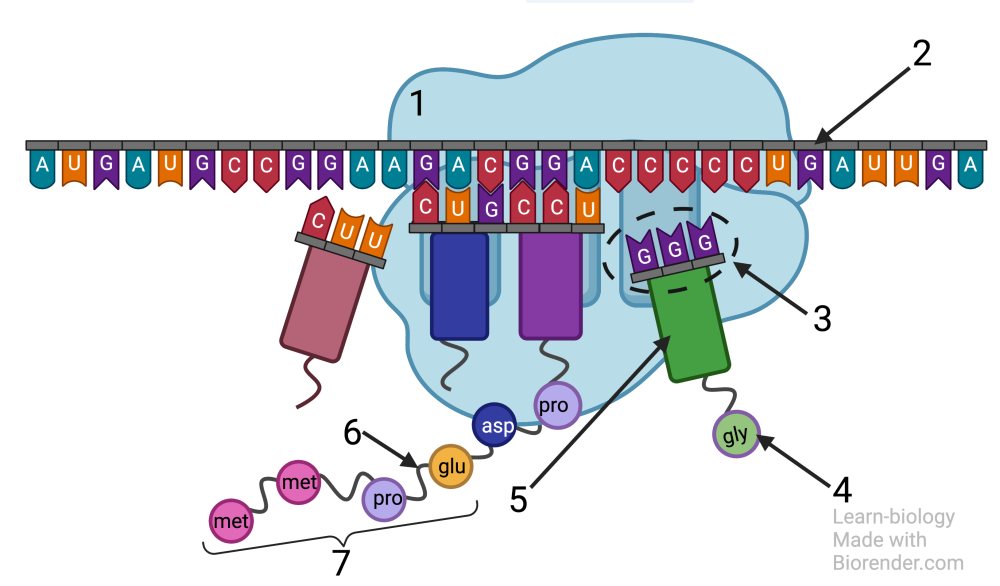
There are about 45 distinct tRNAs. Each one has a distinct anti-codon. Since there are only 20 amino acids, several of the tRNAs carry the same amino acid (which is the basis for the fact that many codons are synonymous, coding for the same amino acid).
4c. Ribosomes are general-purpose protein factories
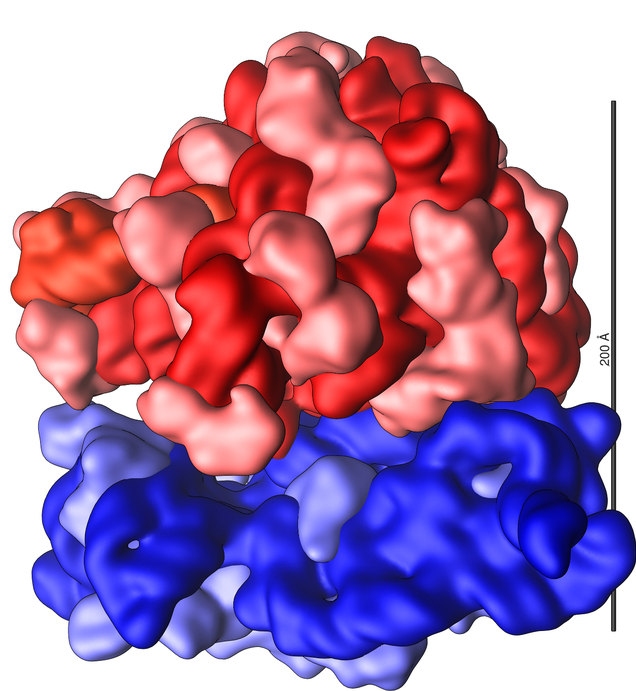
Ribosomes are made of ribosomal RNA (rRNA), along with dozens of proteins.
In terms of function, ribosomes are protein factories capable of reading mRNA instructions and, with the help of tRNAs, assembling any protein the cell needs to make.
In prokaryotic cells, ribosomes are found floating freely in the cytoplasm. In eukaryotic cells, they can be in the cytoplasm, or attached to the endoplasmic reticulum (creating the rough E.R.). Chloroplasts and mitochondria also have their own ribosomes, another remnant of their evolutionary origins and once free-living bacterial cells.
The ribosome is made of two subunits that attach during the process of protein 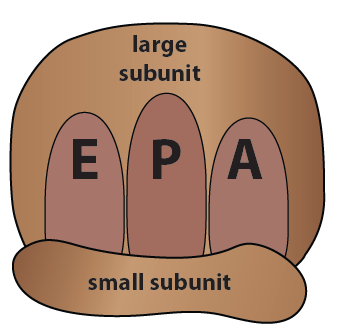 synthesis, then detach after the process is finished. In the image above left, the small subunit is shown in blue, and the large subunit in red. RNA is shown in dark blue and dark red. The lighter shades of blue and red represent proteins.
synthesis, then detach after the process is finished. In the image above left, the small subunit is shown in blue, and the large subunit in red. RNA is shown in dark blue and dark red. The lighter shades of blue and red represent proteins.
In the explanation of the protein synthesis that follows, we’re going to use the image on the right. “E”, “P,” and “A” are binding sites for tRNAs, as we’ll explain below.
5. An overview of translation
During translation, ribosomes “read” the mRNA, translating the sequence of codons in the mRNA into a sequence of amino acids in a protein.
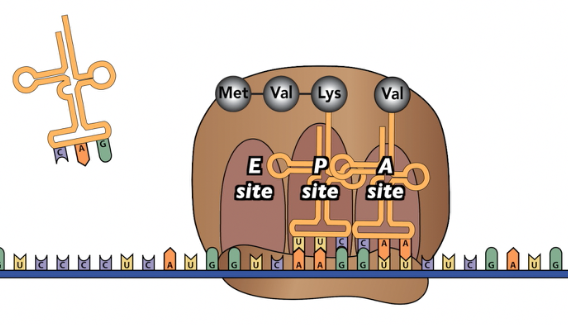
The diagram above shows translation in progress. You’ll watch an animation of this process in a moment, but, for now, notice these key aspects of the process.
- The ribosome is wrapped around the mRNA in a way that exposes the mRNA codons to the cytoplasm.
- The tRNA in the “P” site is holding on to the growing polypeptide, which now consists of three amino acids.
- The tRNA in the “A” site has just entered the ribosome. It’s temporarily held in place by hydrogen bonds between the tRNA’s anticodon and the mRNA’s codon. Imagine that before this tRNA became temporarily stuck, dozens of other tRNAs might have bumped into the “A” site. But because their anti-codon didn’t complement the mRNA codon, each one bounced away.
- What’s the next move? With the correct tRNA in place, the ribosome acts like an enzyme and binds the amino acid in the “A” site to the polypeptide in the “P” site.
Here’s a summary of the three ribosomal binding sites
- The “E” site is the exit site. After a tRNA has “donated” its amino acid to the growing polypeptide, it moves to the E site, and then leaves the ribosome.
- The “P” site is where the ribosome holds onto the growing protein. It’s formally known as the peptidyl-tRNA site.
- The “A” site is where new tRNAs enter the ribosome. Its formal name is the “amino-acyl tRNA site”
6. Protein Synthesis Music Video
Pretty much everything you need to know about protein synthesis at the AP Biology Level is found in my protein synthesis music video. Strap yourself in, because my music partner Max Cowan gives this a heavy metal sound. If you’re in a classroom or a computer lab with other students within earshot, please use your headphones/earbuds.
7. Protein Synthesis, Step-by-Step
What you just watched in the video came by pretty fast. This section explains the same material.
7.1 Initiation
The initiation phase involves the initial binding of the ribosome around the start codon in mRNA. Once the mRNA has been transcribed and has left the nucleus…
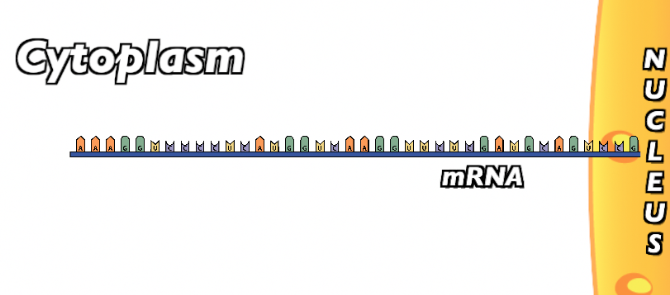
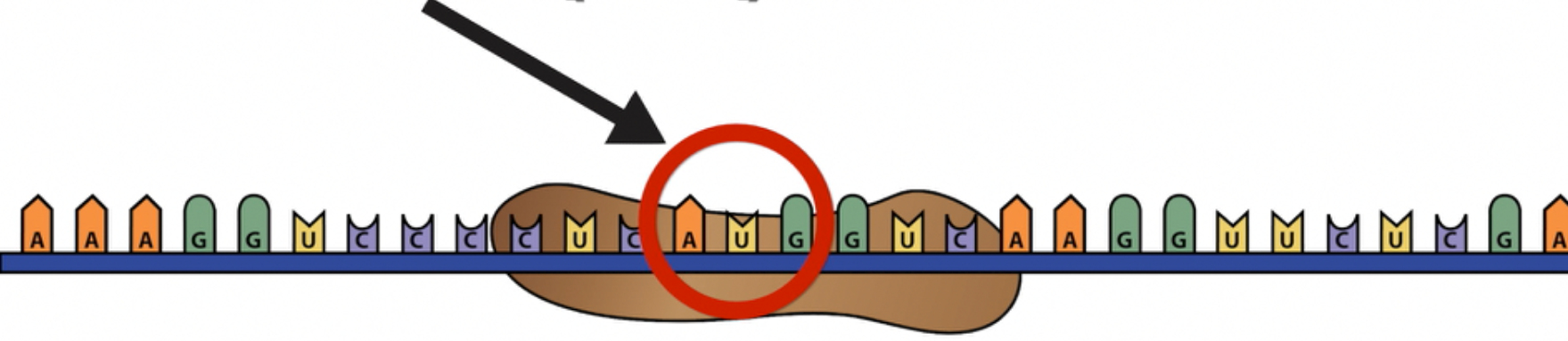
…The small subunit of the ribosome “finds” the start codon AUG.
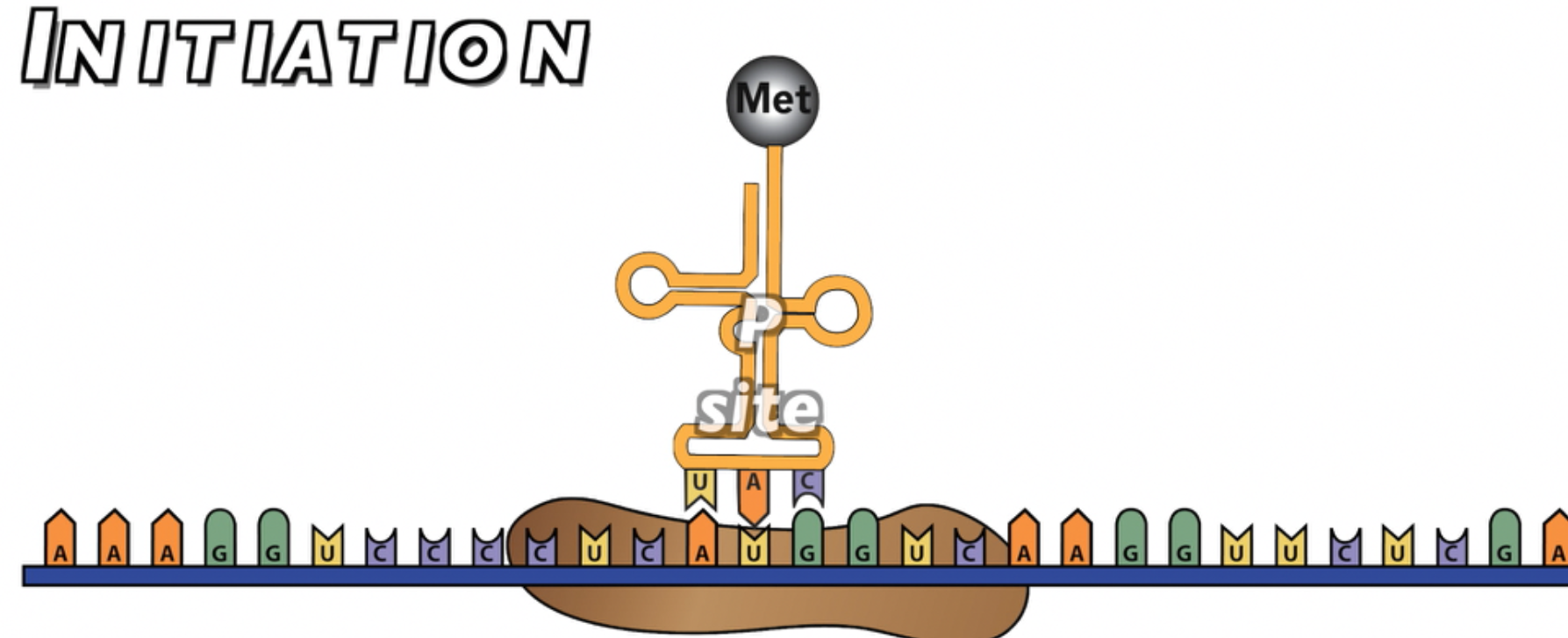
Once in place, the first tRNA can bind. This tRNA binds with the start codon because it has an anti-codon (UAC) that complements the start codon. This first amino acid carries the first amino acid methionine (abbreviated as “Met”).
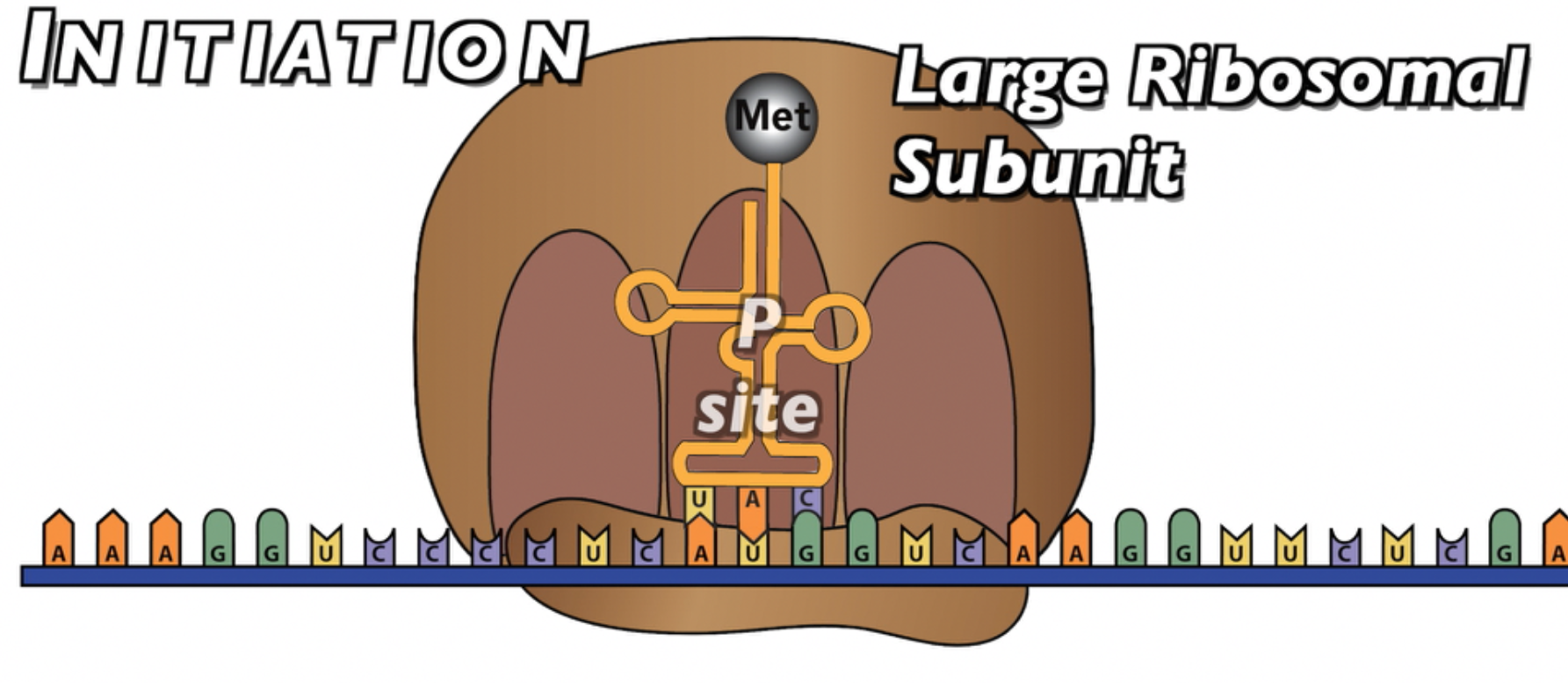
With the first tRNA in place, the large subunit of the ribosome can bind with the small subunit, ending the initiation phase. Note that this first amino acid is now located in the ribosome’s “P site,” the middle of the three binding site.
7.2 Elongation
During the elongation phase, the ribosome builds the polypeptide. Elongation begins as the next tRNA, carrying the next amino acid, enters the ribosome’s “A” site. In the example below, the second tRNA binds with the second codon (GUC) because it has a complementary anti-codon (CAG). As described above, this second tRNA, like the 1st one, is holding on to the mRNA through hydrogen bonds between the codon and anti-codon.
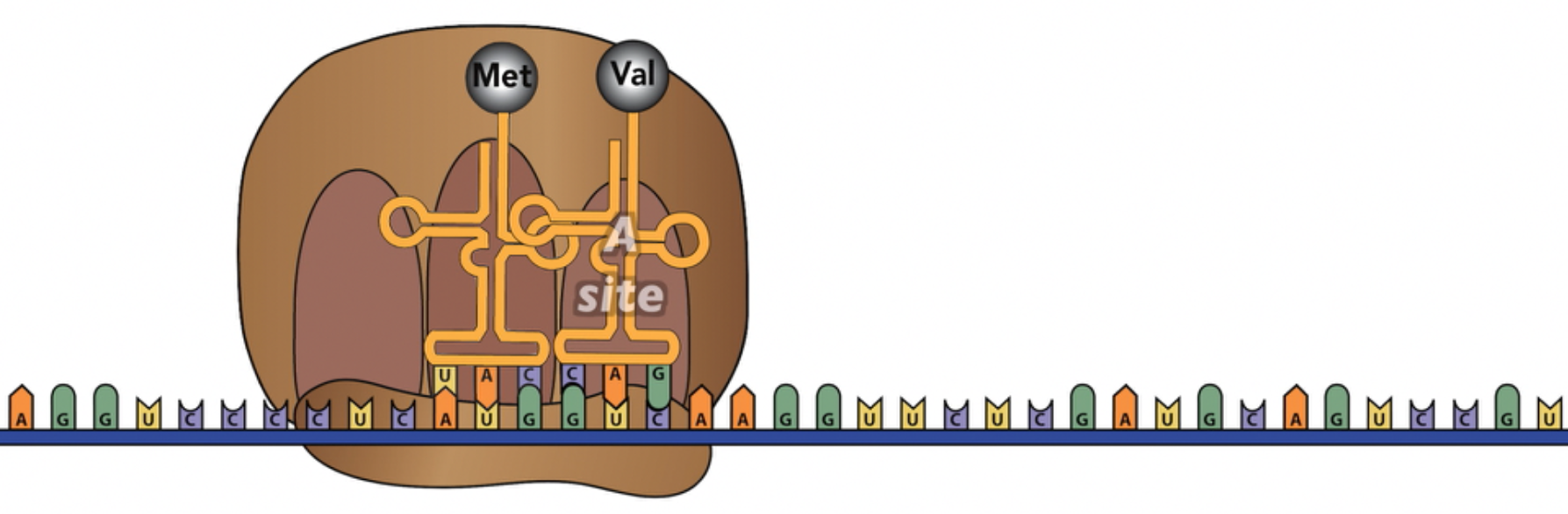
Once the second tRNA is in place, the ribosome acts as an enzyme, catalyzing a peptide bond between the first amino acid (Met) and the second amino acid (Val). At the same time, the bond between Met and the 1st tRNA is broken, so the resulting dipeptide is hanging on to the tRNA that’s in the A site.
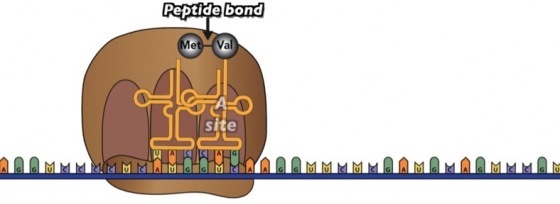
Next comes translocation. The ribosome moves one codon over. Now the A site is empty, and the dipeptide is attached to the tRNA at the P site.
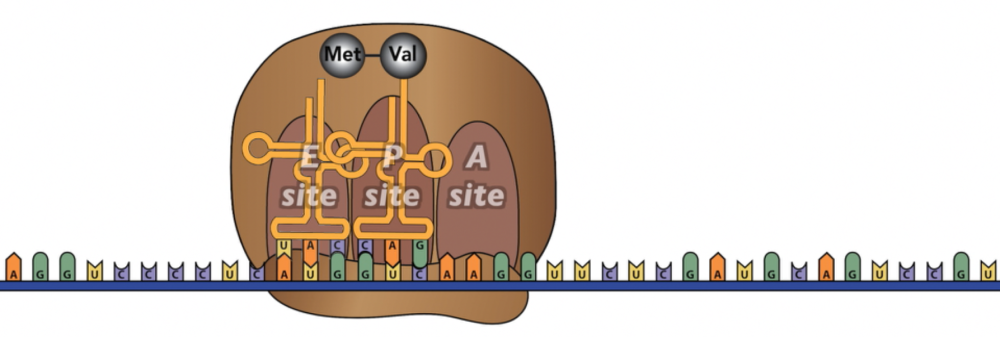
The tRNA in the E site exits.
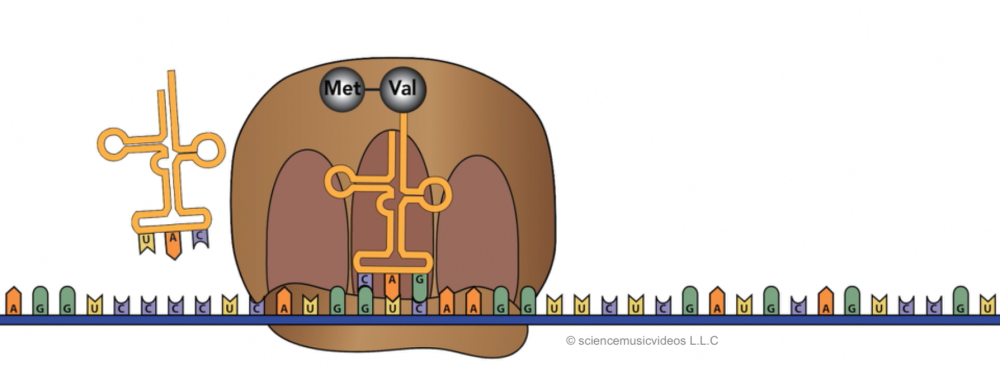
With the A site empty, another tRNA, carrying the next amino acid, can bind.
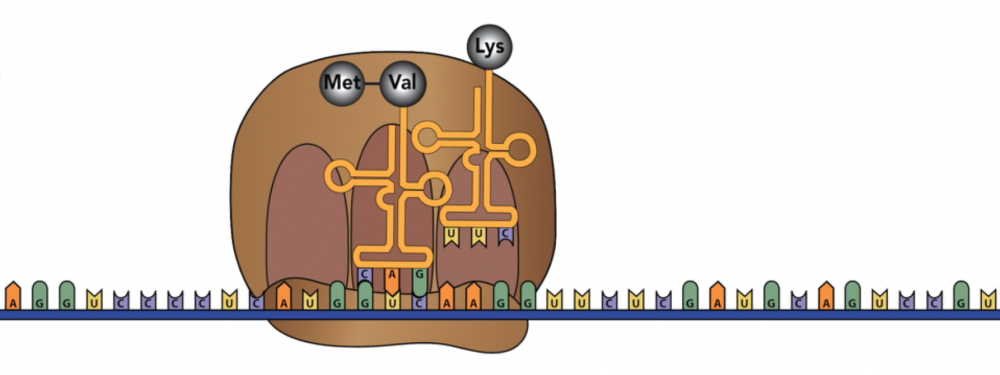
Once a new tRNA is in place at the A site, the ribosome will catalyze a second peptide bond between the new amino acid (in this case, Lysine) with the dipeptide that’s connected to the A site tRNA.
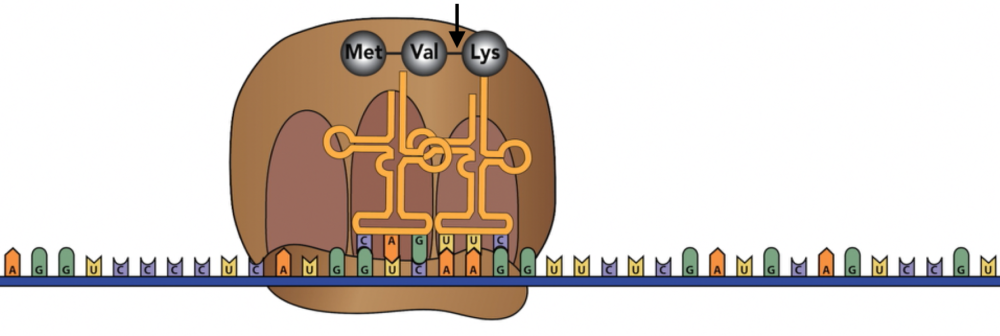
This is followed by successive rounds of 1) translocation, 2) new tRNAs bringing new amino acids to the “A” site, 3) empty tRNAs leaving the “E” site, and 4) peptide bond formation between the newly arrived amino acid and the growing polypeptide change. This will continue for as long as the polypeptide is (which can be hundreds or even thousands of amino acids).
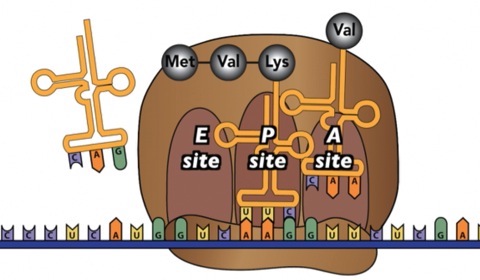
7.3 Termination
At a certain point, the ribosome will arrive at a stop codon.
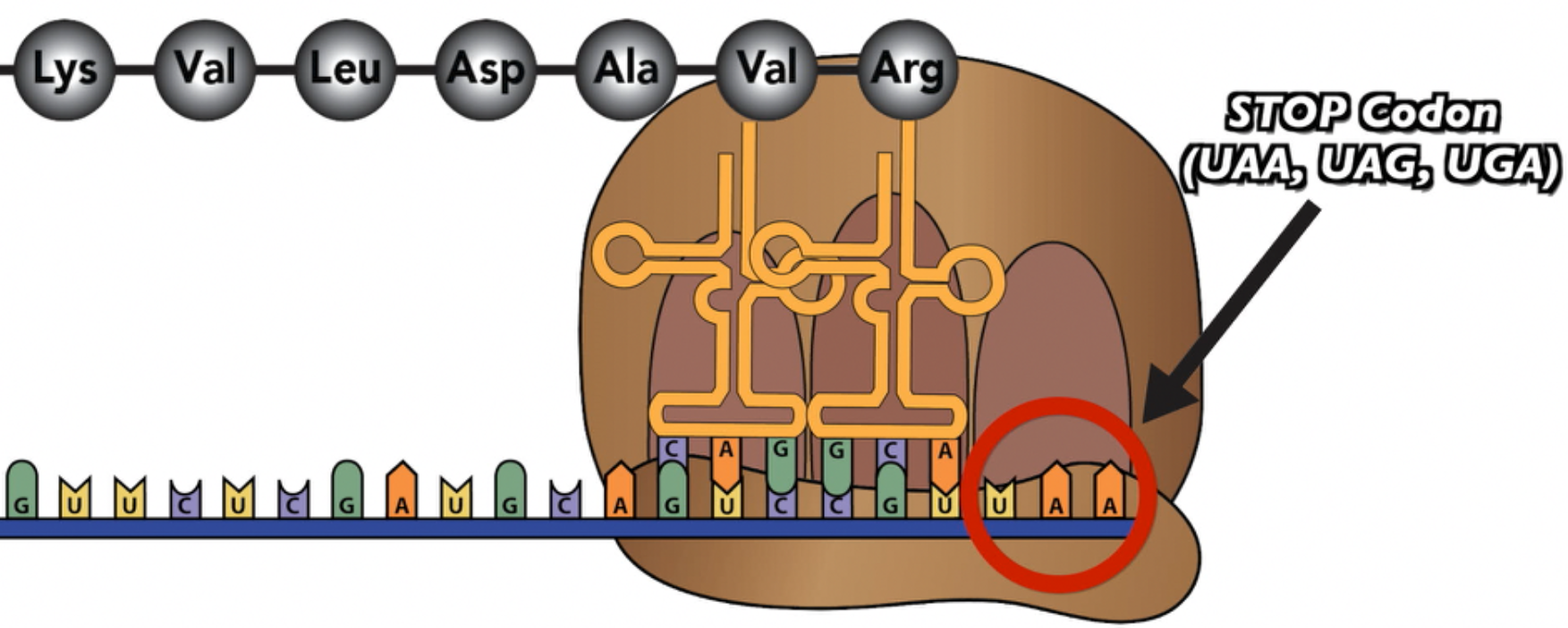
The stop codon, rather than coding for an amino acid, codes for a release factor.
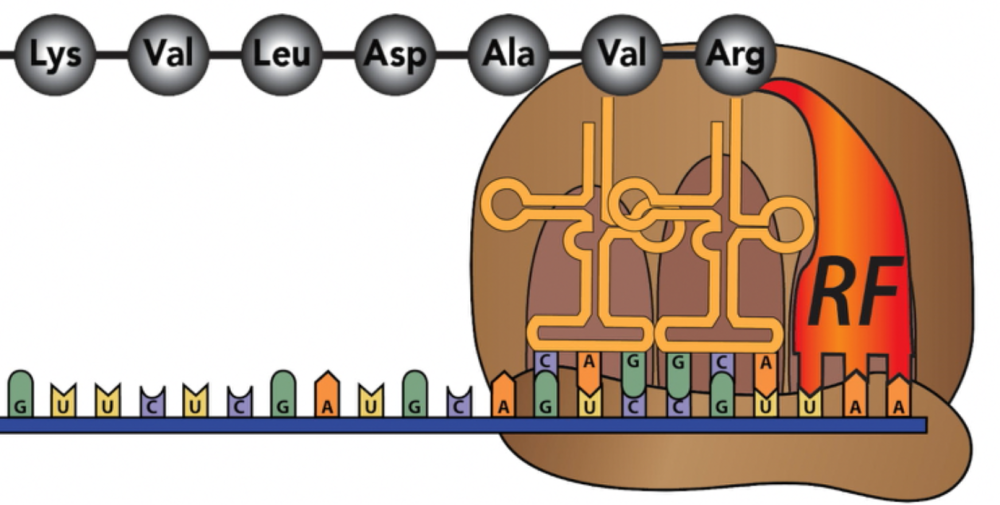
Once the release factor binds, the entire ribosome/mRNA/tRNA/polypeptide complex falls apart.
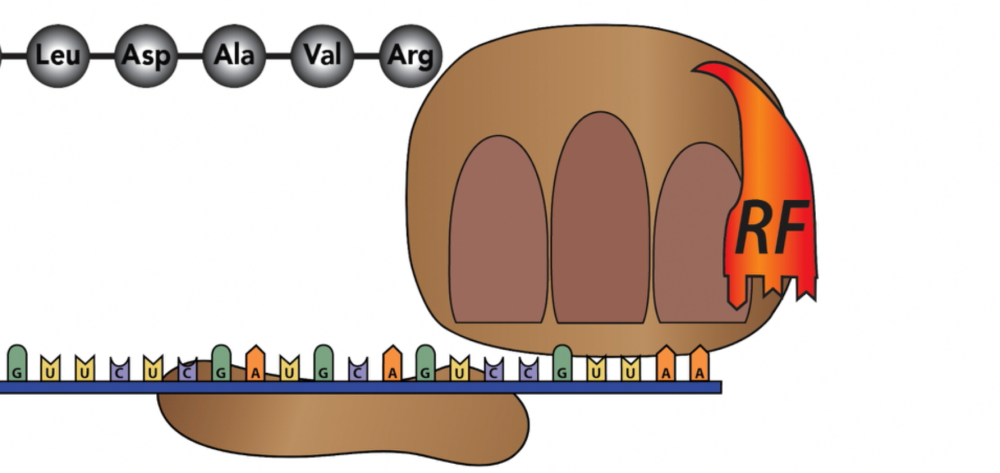
The polypeptide folds up into its 3-D functional shape as a protein (a process that has been occurring throughout the entire protein synthesis process). The small and large subunits go back into the cytoplasm where they’ll be recruited into the synthesis of another protein. The mRNA will probably be hydrolyzed back into RNA monomers (but might also be read again by other ribosomes, especially in prokaryotes).
8. Protein Synthesis Flashcards
[qdeck style=”min-height: 450px width: 650px !important;” bold_text=”false” qrecord_id=”sciencemusicvideosMeister1961-Protein Synthesis Flashcards (v2.0)”]
[h]Protein Synthesis Flashcards
[i]
[q]Describe the function and structure of the ribosome?
[a]Ribosomes are protein factories capable of reading mRNA instructions and, with the help of tRNAs, assembling any protein the cell needs to make.
Ribosomes are composed of both RNA and protein. They have a small subunit and a large subunit. They also have three tRNA binding sites (which are discussed in another card).
[q junit=”6.Gene_Expression_and_Regulation” topic=”6.3.Transcription_and_RNA_Processing”] Describe the function of mRNA. Include the concept of a “codon” in your response.
[a]mRNA (messenger RNA) brings instructions from DNA to ribosomes, which translate mRNA into protein. mRNA’s information is encoded in 3 base sequences called codons, each of which codes for an amino acid, a start codon, or a stop codon.
[q unit=”6.Gene_Expression_and_Regulation” topic=”6.3.Transcription_and_RNA_Processing”] Describe the function of tRNA, and describe a tRNA’s key parts.
[a]tRNAs (transfer RNAs) bring specific amino acids to ribosomes, allowing the ribosomes to bind amino acids together into polypeptides, following the sequence laid down by mRNA. tRNAs have a region called an anticodon: three RNA nucleotides that complement (and bind with) complementary codons on mRNA (which is the basis of the information transfer between RNA and protein). Another key region is the amino acid binding site, to which a specific amino acid gets attached.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|18fecacca9510″ question_number=”215″ topic=”6.3.Transcription_and_RNA_Processing”] Describe the function of rRNA
[a]rRNA (ribosomal RNA), along with protein, makes up the ribosome. The catalytic part of ribosomes that binds amino acids together during protein synthesis is made of rRNA.
[q unit=”6.Gene_Expression_and_Regulation” topic=”6.3.Transcription_and_RNA_Processing”] Explain the overall flow of genetic information of cells.
[a] The basic flow of information in cells is captured in the central dogma of molecular genetics: DNA makes RNA makes protein. More specifically, information flows from a sequence of DNA triplets (3 bases in DNA) to mRNA codons to amino acids.
[q unit=”6.Gene_Expression_and_Regulation” topic=”6.4.Translation”] What is the first phase of translation called? Describe how the process begins.
[a] Translation begins with initiation. The small subunit of the ribosome binds with mRNA, and then slides along the mRNA until it reaches the start codon, AUG. The small subunit pauses, waiting until a tRNA with a matching anticodon and carrying the amino acid methionine binds with the start codon. This sets the stage for the large subunit to bind with the small subunit, ending the initiation phase.
[q unit=”6.Gene_Expression_and_Regulation” topic=”6.4.Translation”] A ribosome has 3 binding sites. What are they, and what do they do?
[a] The ribosome has 3 binding sites, E, P, and A. The “A” site is where the new tRNA, carrying a new amino acid, binds with the mRNA being read by a ribosome. The “P” site is where the tRNA that’s attached to the growing polypeptide binds with the mRNA. The “E” site is where the tRNA that’s about to leave the ribosome is bound to mRNA.
[q unit=”6.Gene_Expression_and_Regulation” topic=”6.4.Translation”] Describe the elongation phase of protein synthesis.
[a] During elongation, the ribosome waits until a tRNA with an anticodon that complements the A site codon binds, bringing in a new amino acid. Once that amino acid is in place, the ribosome catalyzes a peptide bond between the P site and A site amino acids. Next, the ribosome translocates, moving over by one codon. The tRNA in the E site breaks away, leaving its amino acid behind. This leaves a tRNA in the P site, and the A site empty. Another tRNA with an anticodon matching the next codon binds at the A site, and elongation continues, codon after codon.
[q unit=”6.Gene_Expression_and_Regulation” topic=”6.4.Translation”] Describe the termination phase of protein synthesis.
[a] During termination, the ribosome reaches a stop codon. The stop codon has no corresponding amino acid. Instead of another tRNA, a protein called a release factor enters the A site. This causes the polypeptide to dissociate from the last tRNA. The ribosome itself dissociates, breaking into its large and small subunits. Depending on the cell’s needs, the mRNA might be broken down, or it might be translated again.
[q unit=”6.Gene_Expression_and_Regulation” topic=”6.4.Translation”] The genetic code is said to be universal, specific, and redundant. Explain what the genetic code is, and why it has these properties.
[a] The genetic code specifies which three-letter codon sequence in mRNA is translated into which amino acid. It’s universal in that it’s used by all organisms in all three domains of life (bacteria, archaea, and eukarya). The code is nearly identical throughout nature (though there are a few differences where, for example, a stop codon in one group of organisms is translated into a specific amino acid in others). It’s specific in that each codon codes for only one amino acid. It’s redundant in that there are synonyms, with multiple codons coding for the same amino acid.
[q unit=”6.Gene_Expression_and_Regulation” topic=”6.4.Translation”] What is the central dogma of molecular biology?
[a] The central dogma is the idea that genetic information in living organisms flows from DNA to RNA to Protein. DNA is transcribed into RNA, which is translated into protein.
[q]Identify 1 through 7in the diagram below.
[a]
1. ribosome; 2. mRNA; 3. anticodon; 4. amino acid; 5. tRNA; 6. peptide bond; 7. polypeptide
[q] Where are two locations within a eukaryotic cell where ribosomes can be found?
Importance for the AP exam: High
[a] Free ribosomes (3) float freely in the cytoplasm (5). Bound ribosomes (4) are connected to the membrane of the rough ER (endoplasmic reticulum).
[x][restart]
[/qdeck]
9. Protein Synthesis Interactive Diagrams
[qwiz style=”min-height: 450px !important; width: 650px !important;” qrecord_id=”sciencemusicvideosMeister1961-Protein Synthesis Interactive Diagrams (v2.0)”]
[h] Protein Synthesis Interactive Diagrams]
[q xx=”2″ json=”true” labels = “top” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14f1c826d059c9″ question_number=”1″]
[l]amino acid
[f*] Great!
[fx] No. Please try again.
[l]anticodon
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]mRNA
[f*] Good!
[fx] No. Please try again.
[l]large ribosomal subunit
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]small ribosomal subunit
[f*] Correct!
[fx] No. Please try again.
[l]polypeptide
[f*] Excellent!
[fx] No, that’s not correct. Please try again.
[l]tRNA
[f*] Good!
[fx] No. Please try again.
[q xx=”2″ json=”true” labels = “top” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14f01bbe4479c9″ question_number=”3″]
[l]amino acid
[f*] Correct!
[fx] No. Please try again.
[l]anti-codon
[f*] Correct!
[fx] No. Please try again.
[l]mRNA
[f*] Great!
[fx] No. Please try again.
[l]nucleus
[f*] Great!
[fx] No. Please try again.
[l]protein
[f*] Excellent!
[fx] No. Please try again.
[l]ribosome
[f*] Correct!
[fx] No, that’s not correct. Please try again.
[l]tRNA
[f*] Correct!
[fx] No. Please try again.
[q labels = “top”]
[l]amino acid
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]anticodon
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]mRNA
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]peptide bond
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]polypeptide
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]ribosome
[fx] No. Please try again.
[f*] Good!
[l]tRNA
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[x]
[restart]
[/qwiz]
10. Protein Synthesis/Translation Quiz
[qwiz style=”min-height: 450px !important; width: 650px !important;” qrecord_id=”sciencemusicvideosMeister1961-Protein Synthesis/Translation Quiz (v2.0)”][h]Protein Synthesis/Translation Quiz]
[i]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14efa753f1f1c9″ question_number=”4″]Translation occurs at a cell part called a [hangman]. During translation, a message in the form of [hangman] is translated into a sequence of [hangman] acids.
[c]cmlib3NvbWU=[Qq]
[c]bVJOQQ==[Qq]
[c]YW1pbm8=[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14ee51111e05c9″ question_number=”7″]In the diagram below, 1 represents a(n) [hangman] [hangman]. 2 is a(n) [hangman]. The entire structure is a(n) [hangman].
[c]YW1pbm8=[Qq]
[c]IGFjaWQ=[Qq]
[c]YW50aWNvZG9u[Qq]
[c]dFJOQQ==[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14ede5f6fb0dc9″ question_number=”8″]In molecular genetics, “translation” is synonymous with [hangman][hangman]. During translation, three mRNA nucleotides, a unit called a [hangman], are translated into one amino acid.
[c]cHJvdGVpbg==[Qq]
[c]c3ludGhlc2lz[Qq]
[c]Y29kb24=[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14eac05ee8e5c9″ question_number=”15″]In the diagram below, the arrow represents a [hangman] bond connecting two [hangman] [hangman]
[c]cGVwdGlkZQ==[Qq]
[c]YW1pbm8=[Qq]
[c]YWNpZHM=[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14e978145c51c9″ question_number=”18″]The idea that DNA makes RNA makes protein is known as the [hangman] [hangman] of molecular genetics.
[c]Y2VudHJhbA==[Qq]
[c]ZG9nbWE=
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14e5f55a6e89c9″ question_number=”26″]In mRNA, [hangman] RNA nucleotides code for one amino acid (spell out a number).
[c]dGhyZWU=
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14e4a16ba681c9″ question_number=”29″]If a protein was made of 20 amino acids, how many RNA nucleotides would need to code for it? (type out the number)
[hangman]
[c]c2l4dHk=
Cg==Cg==[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14e3c1e73101c9″ question_number=”31″]The structure below is a [hangman]. When it folds up into a specific, functional, 3-dimensional shape, it’s best referred to as a [hangman].
[c]cG9seXBlcHRpZGU=[Qq]
[c]cHJvdGVpbg==[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14e18523c3e9c9″ question_number=”36″]After the small subunit of the ribosome reaches the start codon AUG, what happens next?
[c]QSB0Uk5BIHdpdGjCoGFudGljb2RvbiBBVUcgZm9ybXMgYSBoeWRyb2dlbiBib25kwqB3aXRoIHRoZSBzdGFydCBjb2RvbiBBVUc=[Qq]
[c]QSB0Uk5BIHdpdGggYW50aWNvZG9uIFVBQyBmb3JtcyBhIGh5ZH JvZ2VuIGJvbmTCoHdpdGggdGhlIHN0YXJ0IGNvZG9uIEFVRw==[Qq]
[c]QcKgdFJOQSB3aXRoIGFudGljb2RvbiBVQUMgZm9ybXMgYSBwZXB0aWRlIGJvbmQgd2l0aCB0aGUgc3RhcnQgY29kb24gQVVHLg==[Qq]
[f]Tm8uwqBGb3IgdGhlIHRSTkEgdG8gYmluZCB3aXRoIHRoZSBzdGFydCBjb2RvbiBBVUcsIGl0IG5lZWRzIHRvIGhhdmUgYW4gYW50aWNvZG9uIHRoYXQmIzgyMTc7cyBjb21wbGVtZW50YXJ5IHRvIEFVRywgbm90IGlkZW50aWNhbCB0byBpdC4gV2hhdCYjODIxNztzIG5lZWRlZCBpcyBzaG93biBiZWxvdy4=
Cg==[Qq]
[f]WWVzLsKgRm9yIHRoZSB0Uk5BIHRvIGJpbmQgd2l0aCB0aGUgc3RhcnQgY29kb24gQVVHLCBpdCBuZWVkcyB0byBoYXZlIGFuIGFudGljb2RvbiB0aGF0JiM4MjE3O3MgY29tcGxlbWVudGFyeSB0byBBVUcuIFRoYXQgYW50aWNvZG9uIGlzIFVBQywgYXMgc2hvd24gYmVsb3cu
Cg==[Qq]
[f]Tm8uIFRoZSBib25kcyBiZXR3ZWVuIGFudGljb2RvbiBhbmQgY29kb24gYXJlIGh5ZHJvZ2VuIGJvbmRzLiBQZXB0aWRlIGJvbmRzIG9ubHkgZm9ybSBiZXR3ZWVuIGFkamFjZW50IGFtaW5vIGFjaWRzIGluIHBvbHlwZXB0aWRlcy7CoA==
Cg==[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14dfb1266de5c9″ question_number=”40″]After the small subunit of the ribosome binds with the leading edge of the mRNA, what happens next?
[c]QSB0Uk5BIGJpbmRzIHdpdGggdGhlIGZpcnN0IHNldCBvZiAzIGJhc2VzIG9uIHRoZSBtUk5BLCBiZWdpbm5pbmcgdGhlIHRyYW5zbGF0aW9uIHByb2Nlc3Mu[Qq]
[c]VGhlIGxhcmdlIHN1YnVuaXQgb2YgdGhlIHJpYm9zb21lIGJpbmRzIHdpdGggdGhlIHNtYWxsIHN1YnVuaXQsIGNvbXBsZXRpbmcgdGhlIHJpYm9zb21lLg==[Qq]
[c]QSBwZXB0aWRlIGJvbmQgZm9ybXMgYmV0d2VlbiB0aGUgc21hbGwgc3VidW5pdCBvZiB0aGUgcmlib3NvbWUgYW5kIHRoZSBtUk5B[Qq]
[c]VGhlIHNtYWxsIHN1YnVuaXQgb2YgdGhlIHJpYm9zb21lIHNsaWRlcyBhbG9uZyB0 aGUgbVJOQSB1bnRpbCBpdCByZWFjaGVzIHRoZSBzdGFydCBjb2RvbiwgQVVHLg==[Qq]
[f]Tm8uIHRSTkFzIHdpbGwgb25seSBzdGFydCBiaW5kaW5nIG9uY2UgdGhlIHJpYm9zb21lIHJlYWNoZXMgdGhlIHN0YXJ0IGNvZG9uIEFVRy4gSW4gb3RoZXIgd29yZHMsIHdoYXQmIzgyMTc7cyBiZWxvdyBoYXMgdG8gaGFwcGVuIGZpcnN0Lg==
Cg==[Qq]
[f]Tm8uwqBUaGUgbGFyZ2Ugc3VidW5pdCB3aWxsIG9ubHkgYmluZCBvbmNlIHRoZSBmaXJzdCB0Uk5BIGlzIGluIHBsYWNlLg==
Cg==And that won’t happen until the small subunit reaches the start codon, AUG.
[f]Tm8uIFRoZSBtYWluIHByb2JsZW0gd2l0aCB0aGlzIHJlc3BvbnNlIGlzIHRoYXQgcGVwdGlkZSBib25kc8Kgb25seSBmb3JtIGJldHdlZW4gYWRqYWNlbnQgYW1pbm8gYWNpZHMgaW4gYSBwb2x5cGVwdGlkZS4gVGhlIG5leHQgc3RlcCBpcyBmb3IgdGhlIHNtYWxsIHN1YnVuaXQgb2YgdGhlIHJpYm9zb21lIHRvIHNsaWRlIGFsb25nIHRvIGEgc3BlY2lmaWMgcGxhY2Ugb24gdGhlIG1STkEgdGhhdCBzdGFydHMgdGhlIHByb2JsZW0u[Qq]
[f]WWVzLsKgQWZ0ZXIgYmluZGluZyB3aXRoIHRoZSBsZWFkaW5nIGVkZ2Ugb2YgdGhlIG1STkEsIHRoZSBzbWFsbCBzdWJ1bml0IG9mIHRoZSByaWJvc29tZSBzbGlkZXMgYWxvbmcgdGhlIG1STkEgdW50aWwgaXQgcmVhY2hlcyB0aGUgc3RhcnQgY29kb24sIEFVRy4=
Cg==Cg==[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14df2573a475c9″ question_number=”41″]The proteins produced by a cell are specified by the
[c]c3VnYXIgYW5kIHBob3NwaGF0ZSBzZXF1ZW5jZSBpbiB0aGUgRE5BIG1vbGVjdWxlLg==[Qq]
[c]bnVtYmVyIG9mIHJpYm9zb21lcyBpbiB0aGUgY2VsbC4=[Qq]
[c]bnVjbGVvdGlkZSBzZXF1ZW5jZS BpbiBhIEROQSBtb2xlY3VsZS4=[Qq]
[c]bnVtYmVyIG9mIG1pdG9jaG9uZHJpYSBpbiB0aGUgY2VsbC4=[Qq]
[f]Tm8uIFRoZSBzdWdhciBhbmQgcGhvc3BoYXRlIGluIEROQSBwbGF5IGEgc3RydWN0dXJhbCByb2xlLiBUaGluayBvZiBob3cgcHJvdGVpbnMgYXJlIGNvZGVkIGZvciwgYW5kIHlvdeKAmWxsIGhhdmUgeW91ciBhbnN3ZXIu[Qq]
[f]Tm8uIFRoZSByaWJvc29tZXMgd2lsbCB0cmFuc2xhdGUgYW55IG1STkEgaW5zdHJ1Y3Rpb25zIHRoYXQgY29tZSBhdCB0aGVtLiBUaGluayBvZiBob3cgcHJvdGVpbnMgYXJlIGNvZGVkIGZvciwgYW5kIHlvdeKAmWxsIGhhdmUgeW91ciBhbnN3ZXIu[Qq]
[f]WWVzLiBBcyB0aGUgY2VudHJhbCBkb2dtYSBvZiBtb2xlY3VsYXIgZ2VuZXRpY3Mgc3RhdGVzLCBETkEgbWFrZXMgUk5BIG1ha2VzIHByb3RlaW4uIFRoZSBETkEgc2VxdWVuY2UgdWx0aW1hdGVseSBkZXRlcm1pbmVzIHRoZSBhbWlubyBhY2lkIHNlcXVlbmNlIHRoYXQgbWFrZXMgdXAgYSBzcGVjaWZpYyBwcm90ZWlu[Qq]
[f]Tm8uIFByb3RlaW4gcHJvZHVjdGlvbiBpcyBub3QgY29ubmVjdGVkIHRvIG1pdG9jaG9uZHJpYSAob3RoZXIgdGhhbiBtaXRvY2hvbmRyaWEgcHJvdmlkZSB0aGUgZW5lcmd5IGZvciB0aGUgcHJvY2VzcyBieSBwcm9kdWNpbmcgQVRQKS4gVGhpbmsgb2YgaG93IHByb3RlaW5zIGFyZSBjb2RlZCBmb3IsIGFuZCB5b3XigJlsbCBoYXZlIHlvdXIgYW5zd2VyLg==
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14de92c4b759c9″ question_number=”42″]The function of tRNA molecules is to
[c]dHJhbnNwb3J0IGFtaW5vIGFjaWRzIH RvIG1STkFzIGF0IHJpYm9zb21lcy4=[Qq]
[c]dHJhbnNwb3J0IGFtaW5vIGFjaWRzIHRvIEROQSBpbiB0aGUgbnVjbGV1cy4=[Qq]
[c]c3ludGhlc2l6ZSBtb3JlIHRyYW5zZmVyIFJOQSBtb2xlY3VsZXMu[Qq]
[c]cHJvdmlkZSBhIHRlbXBsYXRlIGZvciB0aGUgc3ludGhlc2lzIG9mIG1STkEu[Qq]
[f]WWVzLiBUaGUgZnVuY3Rpb24gb2YgdFJOQXMgaXMgdG8gdHJhbnNwb3J0IGFtaW5vIGFjaWRzIHRvIG1STkEgYXQgcmlib3NvbWVzLg==[Qq]
[f]Tm8uIHRSTkFzIGFyZSBpbnZvbHZlZCBpbiBwcm90ZWluIHN5bnRoZXNpcy4gV2hlcmUgaXMgdGhhdCBoYXBwZW5pbmc/[Qq]
[f]Tm8uIFN5bnRoZXNpcyBvZiB0Uk5BIGhhcHBlbnMgZHVyaW5nIHRyYW5zY3JpcHRpb24sIGFuZCBpdOKAmXMgY2FycmllZCBvdXQgYnkgUk5BIHBvbHltZXJhc2UgKGFsb25nIHdpdGggYSB0ZWFtIG9mIG90aGVyIGVuenltZXMgYW5kIHByb3RlaW5zKS4gdFJOQXMgYXJlIGludm9sdmVkIGluIHByb3RlaW4gc3ludGhlc2lzLiBXaGVyZSBpcyB0aGF0IGhhcHBlbmluZz8=[Qq]
[f]Tm8uIEROQSBwcm92aWRlcyB0aGUgdGVtcGxhdGUgZm9yIHRoZSBzeW50aGVzaXMgb2YgbVJOQS4gdFJOQXMgYXJlIGludm9sdmVkIGluIHByb3RlaW4gc3ludGhlc2lzLiBXaGVyZSBpcyB0aGF0IGhhcHBlbmluZz8=
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14dda79c0665c9″ question_number=”44″]What’s the first thing that happens during the initiation of protein synthesis?
[c]QSB0Uk5BIGJpbmRzIHdpdGggdGhlIHNtYWxsIHN1YnVuaXQgb2YgdGhlIHJpYm9zb21lLg==[Qq]
[c]VGhlIHNtYWxsIHN1YnVuaXQgb2YgdGhlIHJpYm9zb21lIGJpbm RzIHdpdGggdGhlIG1STkEmIzgyMTc7cyBsZWFkaW5nIGVkZ2Uu[Qq]
[c]VGhlIGxhcmdlIHN1YnVuaXQgb2YgdGhlIHJpYm9zb21lIGZpbmRzIHRoZSBzdGFydCBjb2RvbiBhbmQgdGhlbiBiaW5kcyB3aXRoIHRoZSBzbWFsbCBzdWJ1bml0[Qq]
[c]VGhlIGxhcmdlIHN1YnVuaXQgb2YgdGhlIHJpYm9zb21lIGJpbmRzIHdpdGggYSByZWxlYXNlIGZhY3RvciwgZW5hYmxpbmcgdGhlIHNtYWxsIHN1YnVuaXQgdG8gYmluZC4=[Qq]
[f]Tm8uIEJlZm9yZSB0aGlzIGhhcHBlbnMsIHRoZSBzbWFsbCBzdWJ1bml0IG5lZWRzIHRvIGJpbmQgd2l0aCB0aGUgbVJOQS4=
Cg==[Qq]
[f]WWVzLiBUcmFuc2xhdGlvbiBiZWdpbnMgd2hlbiB0aGUgc21hbGwgc3VidW5pdCBvZiB0aGUgcmlib3NvbWUgYmluZHMgd2l0aCB0aGUgbVJOQSYjODIxNztzIGxlYWRpbmcgZWRnZS4=
Cg==[Qq]
[f]Tm8uwqBUaGUgbGFyZ2Ugc3VidW5pdCBvZiB0aGUgcmlib3NvbWUgY2FuIG9ubHkgYmluZCB3aXRoIHRoZSBzbWFsbCBzdWJ1bml0IGFmdGVyIHRoZSBzbWFsbCBzdWJ1bml0IGJpbmRzIHdpdGggdGhlIG1STkEsIGFuZCB0aGUgZmlyc3QgdFJOQSBiaW5kcy4=
Cg==Cg==[Qq]Before that, the small subunit has to bind with the leading edge of the mRNA.
[f]Tm8uIFRoZSByZWxlYXNlIGZhY3RvciBpcyBpbnZvbHZlZCB3aXRoIHRoZSB0ZXJtaW5hdGlvbiBvZiBwcm90ZWluIHN5bnRoZXNpcywgbm90IGl0cyBzdGFydC4=
Cg==Cg==[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14dd3331b3ddc9″ question_number=”45″]During protein synthesis, the molecule that binds with an amino acid and positions it on a ribosome is
[c]dFJO QS4=[Qq]
[c]bVJOQS4=[Qq]
[c]RE5BLg==[Qq]
[c]clJOQQ==[Qq]
[f]WWVzLiBEdXJpbmcgcHJvdGVpbiBzeW50aGVzaXMsIHRSTkFzIGF0dGFjaCB0byBzcGVjaWZpYyBhbWlubyBhY2lkcyBhbmQgYnJpbmcgdGhlbSB0byB0aGUgcmlib3NvbWUu[Qq]
[f]Tm8uIG1STkEgaXMgdGhlIGNvZGVkIG1lc3NhZ2UgdGhhdOKAmXMgdHJhbnNsYXRlZCBpbnRvIHByb3RlaW4gYXQgcmlib3NvbWVzLiBXaGljaCBraW5kIG9mIFJOQSBicmluZ3MgdGhlIGFtaW5vIGFjaWRzIHRvIHRoZSByaWJvc29tZT8=[Qq]
[f]Tm8uIFdoaWxlIEROQSB1bHRpbWF0ZWx5IHByb3ZpZGVzIHRoZSByZWNpcGUgZm9yIG1ha2luZyBwcm90ZWlucywgaXTigJlzIG5vdCBpbnZvbHZlZCBpbiBicmluZ2luZyBhbWlubyBhY2lkcyB0byB0aGUgcmlib3NvbWUuIFRoYXQgcm9sZSBpcyBwbGF5ZWQgYnkgb25lIG9mIHRoZSBSTkFzLiBXaGljaCBvbmU/[Qq]
[f]Tm8uIHJSTkEsIGFsb25nIHdpdGggcHJvdGVpbiwgbWFrZXMgdXAgdGhlIHJpYm9zb21lLiBXaGljaCBraW5kIG9mIFJOQSBicmluZ3MgdGhlIGFtaW5vIGFjaWRzIHRvIHRoZSByaWJvc29tZT8=
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14dcc11b6d39c9″ question_number=”46″]Each protein in an organism is coded for by an individual
[c]cmlib3NvbWU=[Qq]
[c]Y2hyb21vc29tZS4=[Qq]
[c]bnVjbGVvdGlkZQ==[Qq]
[c]Z2Vu ZQ==[Qq]
[f]Tm8uIFJpYm9zb21lcyBwbGF5IGEga2V5IHJvbGUgaW4gc3ludGhlc2l6aW5nIHByb3RlaW5zLCBidXQgdGhleeKAmXJlIG5vdCB0aGUgY29kZS4gV2hhdCBkbyB3ZSBjYWxsIGEgc2VxdWVuY2Ugb2YgbnVjbGVvdGlkZXMgdGhhdCBjb2RlcyBmb3IgYSBzcGVjaWZpYyBwcm90ZWluPyBIZXJl4oCZcyBhIGhpbnQ6IGl04oCZcyBzb21ldGhpbmcgeW91IGluaGVyaXQu[Qq]
[f]Tm8uIENocm9tb3NvbWVzIGFyZSBlbm9ybW91cyBtb2xlY3VsZXMgb2YgRE5BIHRoYXQgc3RvcmUgZ2VuZXRpYyBpbmZvcm1hdGlvbiBpbiBjZWxscy4gV2hhdCBkbyB3ZSBjYWxsIGEgc2VxdWVuY2Ugb2YgbnVjbGVvdGlkZXMgdGhhdCBjb2RlcyBmb3IgYSBzcGVjaWZpYyBwcm90ZWluPyBIZXJl4oCZcyBhIGhpbnQ6IGl04oCZcyBzb21ldGhpbmcgeW91IGluaGVyaXQu[Qq]
[f]Tm8uIE51Y2xlb3RpZGVzIGFyZSB0aGUgbW9ub21lcnMgb2YgbnVjbGVpYyBhY2lkcy4gQ29tYmluYXRpb25zIG9mIG51Y2xlb3RpZGVzIGFyZSB3aGF0IGNvZGUgZm9yIHByb3RlaW5zLiBXaGF0IGRvIHdlIGNhbGwgYSBzZXF1ZW5jZSBvZiBudWNsZW90aWRlcyB0aGF0IGNvZGVzIGZvciBhIHNwZWNpZmljIHByb3RlaW4/IEhlcmXigJlzIGEgaGludDogaXTigJlzIHNvbWV0aGluZyB5b3UgaW5oZXJpdC4=[Qq]
[f]WWVzLiBFYWNoIHByb3RlaW4gaXMgY29kZWQgZm9yIGJ5IGFuIGluZGl2aWR1YWwgZ2VuZS4=[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14db63dc75a1c9″ question_number=”49″]After the amino acid that a tRNA brings to a ribosome has been attached to a polypeptide, what happens to the tRNA?
[c]SXQgZm9ybXMgYSBzdWdhci1waG9zcGhhdGUgYm9uZCB3aXRoIHRoZSBuZXh0IHRSTkE=[Qq]
[c]SXQgaXMgZGlnZXN0ZWQgYnkgdGhlIHJpYm9zb21lIHNvIHRoYXQgaXRzIG51Y2xlb3RpZGVzIGNhbiBiZSB1c2VkIHRvIGJ1aWxkIG5ldyBSTkE=[Qq]
[c]SXQgZGV0YWNoZXMgZnJvbSB0aGUgcmlib3NvbWUg YW5kIG1vdmVzIGludG8gdGhlIGN5dG9wbGFzbS4=[Qq]
[f]Tm8uIFRoZXJlIGFyZSB0d28gcHJvYmxlbXMgd2l0aCB0aGlzIGFuc3dlci4gMSkgdFJOQXMgZG9uJiM4MjE3O3QgYm9uZCB3aXRoIG9uZSBhbm90aGVyLiAyKSBTdWdhciBwaG9zcGhhdGUgYm9uZHMgYXJlIHRoZSBib25kcyBiZXR3ZWVuIGFkamFjZW50IG51Y2xlb3RpZGVzIGluIGEgbnVjbGVpYyBhY2lkLg==[Qq]
[f]Tm8uIFJpYm9zb21lcyBkbyBoYXZlIHNvbWUgZW56eW1hdGljIHBvd2VycyAodGhleSBjYW4gY2F0YWx5emUgcGVwdGlkZSBib25kcyBiZXR3ZWVuIGFtaW5vIGFjaWRzKSwgYnV0IHRoZXkgZG9uJiM4MjE3O3QgZGlnZXN0IHRSTkFzLg==[Qq]
[f]WWVzLiBBZnRlciBkcm9wcGluZyBvZmYgdGhlaXIgYW1pbm8gYWNpZHMsIHRSTkFzIGRldGFjaCBmcm9tIHRoZSByaWJvc29tZS4=
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14dae875ff6dc9″ question_number=”50″]Protein has a great potential for variation of structure because
[c]ZGlmZmVyZW50IHR5cGVzIG9mIGFtaW5vIGFjaWRzIG1heSBvY2N1ciBpbiBwYWlycy4=[Qq]
[c]ZmF0dHkgYWNpZHMgbWF5IHZhcnk=[Qq]
[c]bWFueSBhbWlubyBhY2lkcyBtYXkgY29tYm luZSBpbiBhIG51bWJlciBvZiB3YXlzLg==[Qq]
[c]bnVjbGVvdGlkZXMgbWF5IHZhcnku[Qq]
[f]Tm8uIFBhaXJpbmcgdXAgYW1pbm8gYWNpZHMgaXMgbm90IGhvdyBwcm90ZWlucyBhcmUgYnVpbHQu[Qq]
[f]Tm8uIFByb3RlaW5zIGFyZSBjb21wb3NlZCBvZiBhbWlubyBhY2lkcywgbm90IGZhdHR5IGFjaWRzLg==[Qq]
[f]WWVzLiBXaXRoIDIwIGFtaW5vIGFjaWRzIHRoYXQgY2FuIGJlIGNvbWJpbmVkIGluIGFueSBvcmRlciwgaW4gc2VxdWVuY2VzIHRoYXQgY2FuIGJlIHVwIHRvIGh1bmRyZWRzIG9mIGFtaW5vIGFjaWRzIGxvbmcsIHByb3RlaW5zIGNhbiBoYXZlIGFuIGVub3Jtb3VzIHZhcmlldHku[Qq]
[f]Tm8uIFByb3RlaW5zIGFyZSBjb21wb3NlZCBvZiBhbWlubyBhY2lkcywgbm90IG51Y2xlb3RpZGVzLg==
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14da78b3c4adc9″ question_number=”51″]What is the function of DNA molecules in the synthesis of proteins?
[c]RE5BIG1vbGVjdWxlcyBjYXRhbHl6ZSB0aGUgZm9ybWF0aW9uIG9mIHBlcHRpZGUgYm9uZHMu[Qq]
[c]RE5BIG1vbGVjdWxlcyBkZXRlcm1pbmUgdGhlIHNlcXVl bmNlIG9mIGFtaW5vIGFjaWRzIGluIGEgcHJvdGVpbi4=[Qq]
[c]RE5BIG1vbGVjdWxlcyB0cmFuc2ZlciBhbWlubyBhY2lkcyBmcm9tIHRoZSBjeXRvcGxhc20gdG8gdGhlIG51Y2xldXMu[Qq]
[f]Tm8uIENhdGFseXppbmcgcGVwdGlkZSBib25kcyBpcyB0aGUgam9iIG9mIHRoZSByaWJvc29tZS4gRE5B4oCZcyByb2xlIGlzIG11Y2ggbW9yZSBpbmZvcm1hdGlvbmFsPw==[Qq]
[f]WWVzLiBETkEgdWx0aW1hdGVseSBkZXRlcm1pbmVzIHRoZSBzZXF1ZW5jZSBvZiBhbWlubyBhY2lkcyBpbiBhIHByb3RlaW4u[Qq]
[f]Tm8uIFRoZSBtYWpvciBwcm9ibGVtIHdpdGggdGhpcyBjaG9pY2UgaXMgdGhhdCBhbWlubyBhY2lkcyBkbyBnZXQgdHJhbnNmZXJyZWQsIGJ1dCBub3QgdG8gdGhlIG51Y2xldXMuIFRoaW5rIG9mIHRoZSBjZW50cmFsIGRvZ21hIG9mIG1vbGVjdWxhciBnZW5ldGljcyAoRE5BIG1ha2VzIFJOQSBtYWtlcyBwcm90ZWluKSBhbmQgeW914oCZbGwgaGF2ZSB5b3VyIGFuc3dlci4=
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14da01f56641c9″ question_number=”52″]The following sequence-AUGCGAUUAAUAUUGAAUUAG-indicates a(n)
[c]RE5BIG1vbGVjdWxlLg==[Qq]
[c]RE5BIG1vbGVjdWxlIHdpdGggYSBtdXRhdGlvbi4=[Qq]
[c]Uk5BIG1vbG VjdWxlLg==[Qq]
[c]YW1pbm8gYWNpZCBzZXF1ZW5jZS4=[Qq]
[f]Tm8uIEROQSBtb2xlY3VsZXMgYXJlIGNvbXBvc2VkIG9mIEROQSBudWNsZW90aWRlcyB3aXRoIHRoZSBuaXRyb2dlbm91cyBiYXNlcyBBLCBULCBDLCBhbmQgRy4gV2hpY2ggbnVjbGVpYyBhY2lkIGhhcyB0aGUgbml0cm9nZW5vdXMgYmFzZSBVIChmb3IgdXJhY2lsKT8=[Qq]
[f]Tm8uIEEgbXV0YXRpb24gd291bGQgc3Vic3RpdHV0ZSBvbmUgRE5BIG51Y2xlb3RpZGUgZm9yIGFub3RoZXIsIGJ1dCB0aGUgcmVzdWx0aW5nIEROQSBtb2xlY3VsZSB3b3VsZCBzdGlsbCBiZSBjb21wb3NlZCBvZiB0aGUgbml0cm9nZW5vdXMgYmFzZXMgQSwgVCwgQywgYW5kIEcuIFdoaWNoIG51Y2xlaWMgYWNpZCBoYXMgVT8=[Qq]
[f]WWVzLiBSTkEgaXMgY29tcG9zZWQgb2YgUk5BIG51Y2xlb3RpZGVzIHdpdGggdGhlIG5pdHJvZ2Vub3VzIGJhc2VzIEEsIFUsIEMsIGFuZCBHLg==[Qq]
[f]Tm8uIEFuIGFtaW5vIGFjaWQgc2VxdWVuY2UgaXMgY29tcG9zZWQgb2YgYW1pbm8gYWNpZHMuIEVhY2ggYW1pbm8gYWNpZCBpcyB1c3VhbGx5IHJlcHJlc2VudGVkIGJ5IGEgdGhyZWUgbGV0dGVyIGFiYnJldmlhdGlvbnMsIGFuZCBhIHNlcXVlbmNlIG9mIGFtaW5vIGFjaWRzIHdvdWxkIGxvb2sgbGlrZSBHTFktU0VSLVBIRS1MRVUuIFRoZSBBLCBVLCBDLCBHIHNlcXVlbmNlIHNob3duIGFib3ZlIHJlcHJlc2VudHMgYSBudWNsZWljIGFjaWQuIFdoaWNoIG9uZT8=
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14d916ccb54dc9″ question_number=”54″]The bonds between codons and anti-codons are
[c]aHlkcm9nZW 4gYm9uZHM=[Qq]
[c]aW9uaWMgYm9uZHM=[Qq]
[c]c3VnYXItcGhvc3BoYXRlIGJvbmRz[Qq]
[c]cGVwdGlkZSBib25kcw==[Qq]
[f]WWVzLiBUaGUgYm9uZCBiZXR3ZWVuIGEgY29kb24gYW5kIGFuIGFudGktY29kb24gaXMgYSBoeWRyb2dlbiBib25kLg==[Qq]
[f]Tm8uIElvbmljIGJvbmRzIGFyZSBib25kcyB0aGF0IG9jY3VyIGJldHdlZW4gaW9ucywgYW5kIGZ1bGx5IGNoYXJnZWQgcGFydGljbGVzLiBUaGUgYm9uZCBiZXR3ZWVuIHNvZGl1bSBhbmQgY2hsb3JpbmUgaW4gdGFibGUgc2FsdCwgTmFDbCBpcyBhbiBpb25pYyBib25kLg==[Qq]
[f]Tm8uIFN1Z2FyLXBob3NwaGF0ZSBib25kcyBhcmUgdGhlIGJvbmRzIHRoYXQgY29ubmVjdCBhZGphY2VudCBudWNsZW90aWRlcyBpbiBudWNsZWljIGFjaWRzLg==[Qq]
[f]Tm8uICYjODIyMDtQZXB0aWRlIGJvbmQmIzgyMjE7IGlzIHRoZSBuYW1lIGdpdmVuIHRvIHRoZSBib25kIGJldHdlZW4gb25lIGFtaW5vIGFjaWQgYW5kIHRoZSBuZXh0IGluIGEgcG9seXBlcHRpZGUu
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14d8a70a7a8dc9″ question_number=”55″]Which of the following molecules, along with proteins, makes up ribosomes?
[c]dFJOQQ==[Qq]
[c]Uk5BIHBvbHltZXJhc2U=[Qq]
[c]clJO QQ==[Qq]
[c]bVJOQQ==[Qq]
[f]Tm8uIHRSTkEgc3RhbmRzIGZvciDigJx0cmFuc2ZlciBSTkEu4oCdIFdoYXQga2luZCBvZiBSTkEgKGFsb25nIHdpdGggcHJvdGVpbikgbWFrZXMgdXAgcmlib3NvbWVzPw==[Qq]
[f]Tm8uIFJOQSBwb2x5bWVyYXNlIGlzIHRoZSBlbnp5bWUgdGhhdCB0cmFuc2NyaWJlcyBSTkEgZnJvbSBETkEuIFdoYXQga2luZCBvZiBSTkEgKGFsb25nIHdpdGggcHJvdGVpbikgbWFrZXMgdXAgcmlib3NvbWVzPw==[Qq]
[f]WWVzLiByUk5BLCBhbG9uZyB3aXRoIHByb3RlaW4sIG1ha2VzIHVwIHJpYm9zb21lcy4=[Qq]
[f]Tm8uIG1STkEgc3RhbmRzIGZvciDigJxtZXNzZW5nZXIgUk5BLuKAnSBXaGF0IGtpbmQgb2YgUk5BIChhbG9uZyB3aXRoIHByb3RlaW4pIG1ha2VzIHVwIHJpYm9zb21lcz8=[Qq]
[q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14d7b2919a09c9″ question_number=”57″]During transcription
[c]Uk5BIHBvbHltZXJhc2Ug YmluZHMgdG8gRE5BLg==[Qq]
[c]dFJOQSBzIGJyaW5nIGFtaW5vIGFjaWRzIHRvIHRoZSByaWJvc29tZXMu[Qq]
[c]dGhlIHJpYm9zb21lIG1vdmVzIGFsb25nIHRoZSBtUk5BIG9uZSBjb2RvbiBhdCBhIHRpbWUu[Qq]
[c]aGVsaWNhc2UgY2F0YWx5emVzIHRoZSAmIzgyMjA7dW56aXBwaW5nJiM4MjIxOyBvZiBETkEu[Qq]
[f]WWVzLiBEdXJpbmcgdHJhbnNjcmlwdGlvbiwgUk5BIHBvbHltZXJhc2UgYmluZHMgdG8gRE5BIGFuZCBtYWtlcyBSTkEu[Qq]
[f]Tm8uIHRSTkFzIGJyaW5nIGFtaW5vIGFjaWRzIHRvIHRoZSByaWJvc29tZXMgZHVyaW5nIHRyYW5zbGF0aW9uLiBUaGluayBhYm91dCB3aGF04oCZcyBnZXR0aW5nIG1hZGUgZHVyaW5nIHRyYW5zY3JpcHRpb24sIGFuZCB5b3XigJlsbCBoYXZlIHRoZSBhbnN3ZXIu[Qq]
[f]Tm8uIFJpYm9zb21lcyBtb3ZlIGFsb25nIHRoZSBtUk5BIGR1cmluZyB0cmFuc2xhdGlvbi4gVGhpbmsgYWJvdXQgd2hhdOKAmXMgZ2V0dGluZyBtYWRlIGR1cmluZyB0cmFuc2NyaXB0aW9uLCBhbmQgeW914oCZbGwgaGF2ZSB0aGUgYW5zd2VyLg==[Qq]
[f]Tm8uIEhlbGljYXNlIHVuaXppcHMgRE5BIGR1cmluZyByZXBsaWNhdGlvbi4gVGhpbmsgYWJvdXQgd2hhdOKAmXMgZ2V0dGluZyBtYWRlIGR1cmluZyB0cmFuc2NyaXB0aW9uLCBhbmQgeW914oCZbGwgaGF2ZSB0aGUgYW5zd2VyLg==
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14d7372b23d5c9″ question_number=”58″]The bonds connecting the amino acids in a polypeptide are
[c]aHlkcm9nZW4gYm9uZHM=[Qq]
[c]aW9uaWMgYm9uZHM=[Qq]
[c]c3VnYXItcGhvc3BoYXRlIGJvbmRz[Qq]
[c]cGVwdGlkZS Bib25kcw==[Qq]
[f]Tm8uIEh5ZHJvZ2VuIGJvbmRzIGFyZSB3ZWFrIGJvbmRzIHRoYXQgb2NjdXIgYmV0d2VlbiBwb2xhciBtb2xlY3VsZXMsIG9yIGJldHdlZW4gcG9sYXIgcGFydHMgb2YgdGhlIHNhbWUgbW9sZWN1bGUuIFRoZSBib25kIGJldHdlZW4gYWRlbmluZSBhbmQgdGh5bWluZSBpbiBETkEgaXMgYSBoeWRyb2dlbiBib25kLg==[Qq]
[f]Tm8uwqBJb25pYyBib25kcyBhcmUgYm9uZHMgdGhhdCBvY2N1ciBiZXR3ZWVuIGlvbnMsIGZ1bGx5IGNoYXJnZWQgcGFydGljbGVzLiBUaGUgYm9uZCBiZXR3ZWVuIHNvZGl1bSBhbmQgY2hsb3JpbmUgaW4gdGFibGUgc2FsdCwgTmFDbCBpcyBhbiBpb25pYyBib25kLg==[Qq]
[f]Tm8uIFN1Z2FyLXBob3NwaGF0ZSBib25kcyBhcmUgdGhlIGJvbmRzIHRoYXQgY29ubmVjdCBhZGphY2VudCBudWNsZW90aWRlcyBpbiBudWNsZWljIGFjaWRzLg==[Qq]
[f]WWVzLiAmIzgyMjA7UGVwdGlkZSBib25kJiM4MjIxOyBpcyB0aGUgbmFtZSBnaXZlbiB0byB0aGUgYm9uZCBiZXR3ZWVuIG9uZSBhbWlubyBhY2lkIGFuZCB0aGUgbmV4dCBpbiBhIHBvbHlwZXB0aWRlLg==
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14d649ae66fdc9″ question_number=”60″]The movement of a ribosome from one codon to the next down the length of an mRNA strand is called
[c]dHJhbnNsYXRpb24=[Qq]
[c]dHJhbnNjcmlwdGlvbg==[Qq]
[c]dHJhbnNsb2 NhdGlvbg==[Qq]
[f]Tm8uwqBUcmFuc2xhdGlvbiBpcyB0aGUgbmFtZSBmb3IgdGhlIGVudGlyZSBwcm9jZXNzIG9mIHByb3RlaW4gc3ludGhlc2lzLg==[Qq]
[f]Tm8uwqBUcmFuc2NyaXB0aW9uIGlzIHRoZSBtYWtpbmcgb2bCoFJOQSBmcm9tIGEgRE5BIHRlbXBsYXRlLg==[Qq]
[f]WWVzLiBUcmFuc2xvY2F0aW9uIGlzIHRoZSBtb3ZlbWVudCBvZiBhIHJpYm9zb21lIGRvd24gdGhlIGxlbmd0aCBvZiBhbiBtUk5BIHN0cmFuZC4=
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14d5d09bfcadc9″ question_number=”61″]mRNA is responsible for
[c]cHJvZHVjdGlvbiBvZiByaWJvc29tZXMu[Qq]
[c]YnJpbmdpbmcgaW5zdHJ1Y3Rpb2 5zIHRvIHRoZSByaWJvc29tZXMu[Qq]
[c]cmVhZGluZyB0aGUgYW1pbm8gYWNpZCBjb2RlIGZyb20gdFJOQS4=[Qq]
[c]dHJhbnNjcmlwdGlvbiBvZiBETkEgaW50byBSTkEu[Qq]
[c]YnJpbmdpbmcgYW1pbm8gYWNpZHMgdG8gdGhlIHJpYm9zb21lcy4=[Qq]
[f]Tm8uIFByb2R1Y3Rpb24gb2Ygcmlib3NvbWVzIGludm9sdmVzIHByb2R1Y2luZyBSTkEgYW5kIHByb3RlaW4uIFByb2R1Y2luZyBSTkEgaXMgYSBmdW5jdGlvbiBvZiBSTkEgcG9seW1lcmFzZS4gUHJvZHVjaW5nIHByb3RlaW4gaW52b2x2ZXMgbVJOQSwgYnV0IGFsc28gdFJOQXMgYW5kIHJpYm9zb21lcy4gbVJOQSBoYXMgYSBrZXkgam9iIGR1cmluZyBwcm90ZWluIHN5bnRoZXNpcy4gV2hhdCBpcyBpdD8=[Qq]
[f]WWVzLiBtUk5B4oCZcyBmdW5jdGlvbiBpcyB0byBicmluZyBpbnN0cnVjdGlvbiB0byB0aGUgcmlib3NvbWVzLg==[Qq]
[f]Tm8uIFRoaXMgY2hvaWNlIGhhcyBzZXZlcmFsIHByb2JsZW1zLiAxKSBUaGUgY29kZSBpcyBmb3IgcHJvdGVpbnMsIG5vdCBhbWlubyBhY2lkcyAodGhlIG1vbm9tZXJzIG9mIHByb3RlaW4pLiAyKSBUaGUgY29kZSBpcyBvbiB0aGUgbVJOQSwgbm90IHRSTkEgKGFuZCB0aGF04oCZcyBhIGhpbnQgdG8gdGhlIGFuc3dlciB3aGVuIHlvdSBzZWUgdGhpcyBxdWVzdGlvbiBsYXRlciku[Qq]
[f]Tm8uIFRoZSBlbnp5bWUgUk5BIHBvbHltZXJhc2UgdHJhbnNjcmliZXMgRE5BIGludG8gUk5BLiBNZXNzZW5nZXIgUk5BIChtUk5BKSBoYXMgYSBrZXkgam9iIGR1cmluZyBwcm90ZWluIHN5bnRoZXNpcy4gV2hhdCBpcyBpdD8=[Qq]
[f]Tm8uIHRSTkEgaXMgdGhlIG1vbGVjdWxlIHRoYXQgYnJpbmdzIGFtaW5vIGFjaWRzIHRvIHRoZSByaWJvc29tZXMuIE1lc3NlbmdlciBSTkEgKG1STkEpIGhhcyBhIGtleSBqb2IgZHVyaW5nIHByb3RlaW4gc3ludGhlc2lzLiBXaGF0IGlzIGl0Pw==
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14d5508d6eb1c9″ question_number=”62″]Translation is the process in which
[c]RE5BIG1ha2VzIGFuIGV4YWN0IGNvcHkgb2YgaXRzZWxmLg==[Qq]
[c]dGhlIEROQSBzdHJhbmQgdW53aW5kcyBhbmQgdHJhbnNjcmliZXMgaW5mb3JtYXRpb24gdG8gbVJOQS4=[Qq]
[c]aW5mb3JtYXRpb24gaW4gY29kb25zIGlzIHVzZWQgdG 8gYnVpbGQgY2hhaW5zIG9mIGFtaW5vIGFjaWRzLg==[Qq]
[c]Uk5BIGxlYXZlcyB0aGUgbnVjbGV1cyB0aHJvdWdoIG51Y2xlYXIgcG9yZXMu[Qq]
[f]Tm8uIENvcHlpbmcgRE5BIGlzIHJlcGxpY2F0aW9uLiBJbiBtb2xlY3VsYXIgZ2VuZXRpY3MsIHdoYXQgaXMgdHJhbnNsYXRpb24gYSBzeW5vbnltIGZvcj8=[Qq]
[f]Tm8uIFdoZW4gdGhlIEROQSBzdHJhbmQgdW53aW5kcyBhbmQgdHJhbnNjcmliZXMgaW5mb3JtYXRpb24gdG8gbVJOQSwgdGhhdOKAmXMgdHJhbnNjcmlwdGlvbi4gbiBtb2xlY3VsYXIgZ2VuZXRpY3MsIHdoYXQgaXMgdHJhbnNsYXRpb24gYSBzeW5vbnltIGZvcj8=[Qq]
[f]WWVzLiBUcmFuc2xhdGlvbiBpcyB3aGVuIGNlbGxzIHVzZSB0aGUgaW5mb3JtYXRpb24gaW4gUk5BIGNvZG9ucyB0byBidWlsZCBjaGFpbnMgb2YgYW1pbm8gYWNpZHMu[Qq]
[f]Tm8uIFJOQeKAmXMgbW92ZSBpbnRvIHRoZSBjeXRvcGxhc20gaGFwcGVucyBiZWZvcmUgdHJhbnNsYXRpb24gb2NjdXJzLiBXaGF0IGhhcHBlbnMgZHVyaW5nIHRyYW5zbGF0aW9uIGl0c2VsZj8=
Cg==Cg==[Qq][q xx=”2″ json=”true” dataset_id=”Protein_synth_MC, HM, ID (AP-DS)|14d43b7be7b5c9″ question_number=”64″]A ribosome reaches the stop codon on an mRNA. The next thing that will happen is
[c]dGhlIHRSTkFzIGRldGFjaCBmcm9tIHRoZWlyIGJpbmRpbmcgc2l0ZXMu[Qq]
[c]YSByZWxlYXNlIGZhY3RvciBiaW5kcy B3aXRoIHRoZSBzdG9wIGNvZG9uLg==[Qq]
[c]dGhlIHJpYm9zb21lIGZhbGxzIGFwYXJ0[Qq]
[c]dGhlIHBvbHlwZXB0aWRlIGlzIHJlbGVhc2Vk[Qq]
[f]Tm8uwqBCZWZvcmUgdGhlIHRSTkFzIGRldGFjaCwgc29tZXRoaW5nIGhhcyB0byBiaW5kIHdpdGggdGhlIHN0b3AgY29kb24u[Qq]
[f]WWVzLiBBIHJlbGVhc2UgZmFjdG9yIHdpbGwgYmluZCB3aXRoIHRoZSBzdG9wIGNvZG9uLCBsZWFkaW5nIHRoZSBwb2x5cGVwdGlkZSB0byBkZXRhY2ggYW5kIHRoZSBlbnRpcmUgcmlib3NvbWUgYXNzZW1ibHkgc3RydWN0dXJlIHRvIGZhbGwgYXBhcnQu[Qq]
[f]Tm8uwqBCZWZvcmUgdGhlIHJpYm9zb21lIGZhbGxzIGFwYXJ0LCBzb21ldGhpbmcgaGFzIHRvIGJpbmQgd2l0aCB0aGUgc3RvcCBjb2Rvbi4=[Qq]
[f]Tm8uwqBCZWZvcmUgdGhlIHBvbHlwZXB0aWRlIGlzIHJlbGVhc2VkLCBzb21ldGhpbmcgaGFzIHRvIGJpbmQgd2l0aCB0aGUgc3RvcCBjb2Rvbi4=[Qq]
[q]The diagram below represents the end of the [hangman] phase of protein synthesis.
[c]dGVybWluYXRpb24=[Qq]
[q]The diagram below shows the [hangman] phase of protein synthesis.
[c]ZWxvbmdhdGlvbg==[Qq]
[q]Once the tRNA carrying “Val” is in place, the next thing that will happen is
[c]dGhlIHJpYm9zb21lIHdpbGwgdHJhbnNsb2NhdGUgb25lIGNvZG9uIG92ZXIgdG8gdGhlIGNvZG9uICYjODIyMDtVQ0cmIzgyMjE7[Qq]
[c]dGhlIHJpYm9zb21lIHdpbGwgY2F0YWx5emUgYSBwZXB0aWRlIGJvbmQgYmV0 d2VlbiAmIzgyMjA7THlzJiM4MjIxOyBhbmQgJiM4MjIwO1ZhbCYjODIyMTs=[Qq]
[c]VGhlIHJpYm9zb21lIHdpbGwgY2F0YWx5emUgYSBwZXB0aWRlIGJvbmQgYmV0d2VlbiAmIzgyMjA7Q0FBJiM4MjIxOyBhbmQgJiM4MjIwO0dVVSYjODIyMTs=[Qq]
[c]VGhlIHRSTkEgYXQgdGhlIFAgc2l0ZSB3aWxsIGxlYXZlIHRoZSByaWJvc29tZS4=[Qq]
[f]Tm8uIEJlZm9yZSB0cmFuc2xvY2F0aW9uLCB0aGUgcmlib3NvbWUgbmVlZHMgdG8gYXR0YWNoIHRoZSBwb2x5cGVwdGlkZSB0aGF0JiM4MjE3O3MgaGFuZ2luZyBvbiB0byB0aGUgUCBzaXRlIHRSTkEgdG8gdGhlIG5ldyBhbWlubyBhY2lkIHRoYXQgd2lsbCBhcnJpdmUgYXQgdGhlIEEgc2l0ZS4=[Qq]
[f]RXhhY3RseSEgVGhlIHJpYm9zb21lIHdpbGwgY2F0YWx5emUgYSBwZXB0aWRlIGJvbmQgYmV0d2VlbiB0aGUgbmV3bHkgYXJyaXZlZCBhbWlubyBhY2lkIGFuZCB0aGUgcG9seXBlcHRpZGUgdGhhdCYjODIxNztzIGhhbmdpbmcgb24gdGhlIFAgc2l0ZSB0Uk5BLg==[Qq]
[f]Tm8uIFRoZSBib25kIHRoYXQgZm9ybXMgYmV0d2VlbiBhbnRpY29kb24gQ0FBIGFuZCBjb2RvbiBVQ0cgaXMgYSBoeWRyb2dlbiBib25kLg==[Qq]
[f]Tm8uIEJlZm9yZSBleGl0LCB0aGUgcmlib3NvbWUgbmVlZHMgdG8gY2F0YWx5emUgYSBuZXcgcGVwdGlkZSBib25kLCBhbmQgdGhlbiB0cmFuc2xvY2F0ZS4=[Qq]
[x]
[restart]
[/qwiz]
What’s next?
Proceed to Topic 6.4, part 3 (extension): Protein Targeting to the Rough ER
