1. DNA Replication: The Big Picture
DNA Replication means “copying DNA” It involves making two new molecules of “daughter” DNA from one molecule of “parent” DNA.
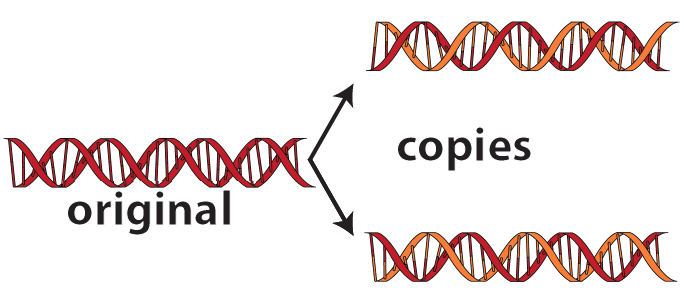
Here’s a big-picture view of what happens during DNA Replication.
- Enzymes (not shown) separate the DNA double-helix into two single strands.
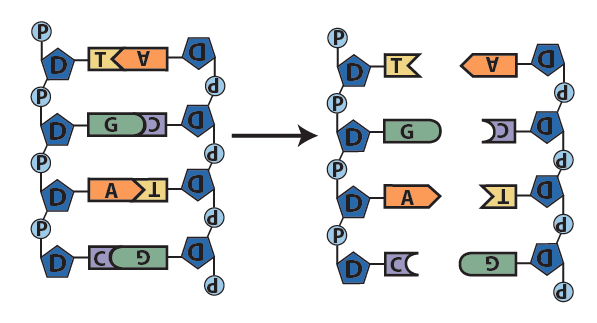
Step 1: the double helix separates into two single strands along part of its length. - With bases exposed, each single strand now serves as a template for the synthesis of a new strand. A template is “a molecule … that serves as a pattern for the generation of another macromolecule” (Meriam-Webster, online).
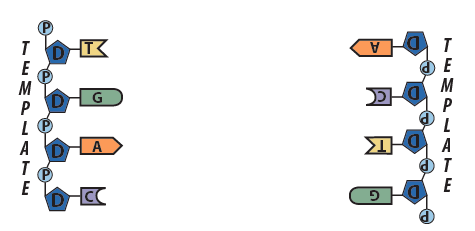
Step 2: Each strand serves as a template for the synthesis of a new strand. - Following the base pairing rules, new complementary nucleotides form hydrogen bonds with nucleotides on the template strands.
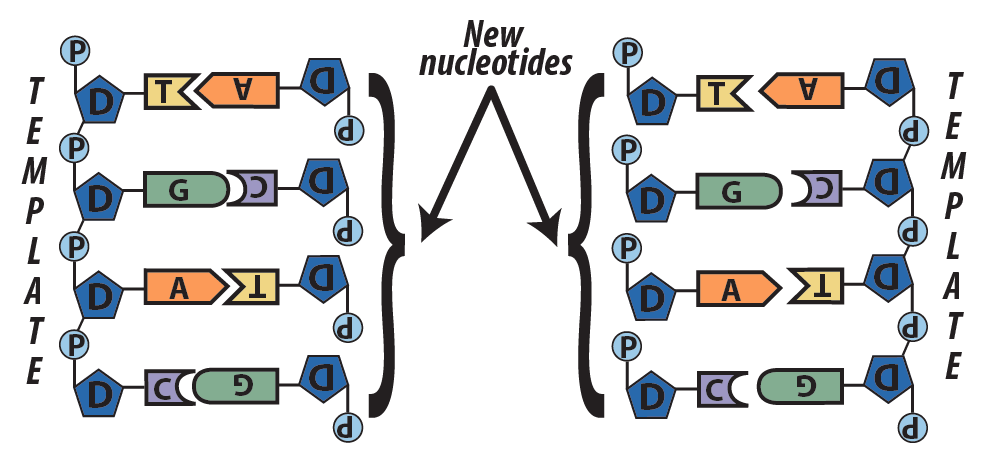
Step 3: new complementary nucleotides bind with the parent strands. - Enzymes create covalent bonds between the sugars and the phosphates of adjacent nucleotides.
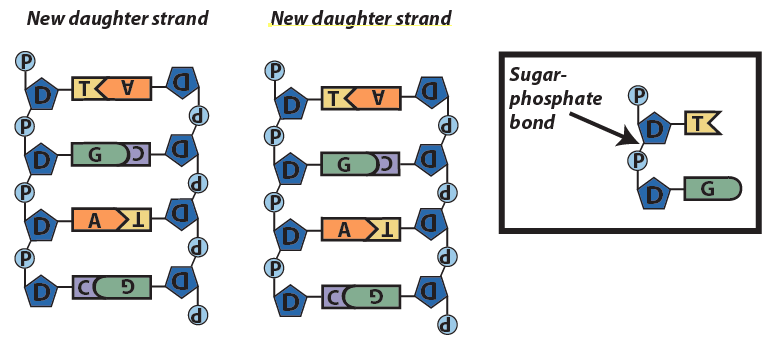
Step 4: Enzymes bind the new strands together with sugar-phosphate bonds.
This model of replication is called semi-conservative. That’s because in each new strand, one strand is old, and one strand is new. The old strand is called the conserved strand because it was conserved from the parent strand.

Here’s the same explanation in verse:
You first unzip the DNA in one or more places,
Breaking hydrogen bonds to separate the bases.
Each resulting single strand serves as a template,
Allowing enzymes to replicate
New strands with complementary bases that match
And through hydrogen bonds, these bases attach
Each nucleotide now bonds to the next
Through a sugar-phosphate bond they connect
Meselsohn and Stahl proved in ‘58 (read about their experiment)
That this is how the double helix replicates
One strand new, the parent strand preserved,
In other words, the whole thing is semi-conserved.
2. Checking Understanding: DNA Replication (the big picture)
[qwiz random = “true” qrecord_id=”sciencemusicvideosMeister1961-DNA Replication, Big Picture (v2.0)”]
[h]DNA replication: The Big Picture
[i]
[q]Copying of DNA is more formally known as DNA [hangman].
[c]cmVwbGljYXRpb24=[Qq]
[q]During replication, each strand acts as a [hangman] for the synthesis of a new strand.
[c]dGVtcGxhdGU=[Qq]
[q]During DNA replication, the new nucleotides that bind with the template strands are [hangman] to the nucleotides on the template strands. These new nucleotides form [hangman] bonds with the template strand nucleotides.
[c]Y29tcGxlbWVudGFyeQ==[Qq]
[c]aHlkcm9nZW4=[Qq]
[q]Whereas the bonds between nitrogenous bases are [hangman] bonds, the bonds between the sugars and phosphates on adjacent nucleotides are [hangman] bonds.
[c]aHlkcm9nZW4=[Qq]
[c]Y292YWxlbnQ=[Qq]
[q]Replication ends as enzymes create covalent bonds between [hangman] and phosphates on adjacent nucleotides.
[c]c3VnYXJz
Cg==[Qq]
[q]The result of replication is two daughter DNA molecules, each of which has one [hangman] strand and one old strand.
[c]bmV3[Qq]
[q]Because replication results in daughter strands that are half new and half old, replication is called [hangman]-[hangman].
[c]c2VtaQ==[Qq]
[c]Y29uc2VydmF0aXZl[Qq]
[/qwiz]
3. How DNA replication occurs in cells
When you look at a model or diagram of DNA and go about the task of replicating it, you can see the bases, think of the base pairing rules, and manipulate the model (pulling it apart, adding new nucleotides) to create daughter strands from the parent strand. How can cells, which can’t see or think, perform this task?
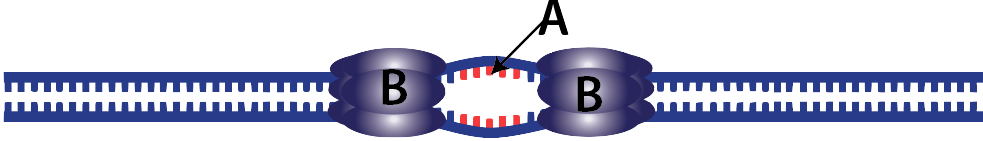
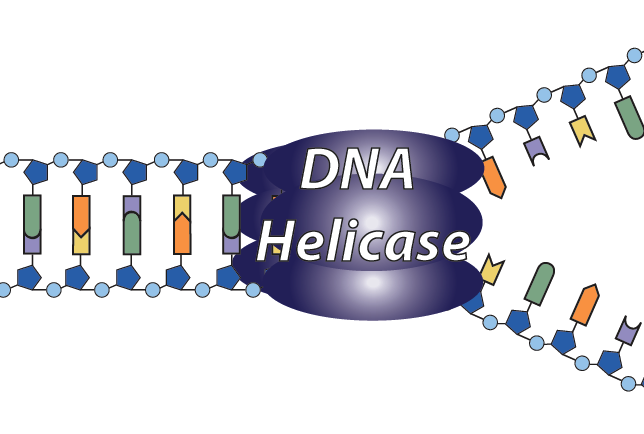
2. The structure that results from the opening of the helix is called a replication fork.
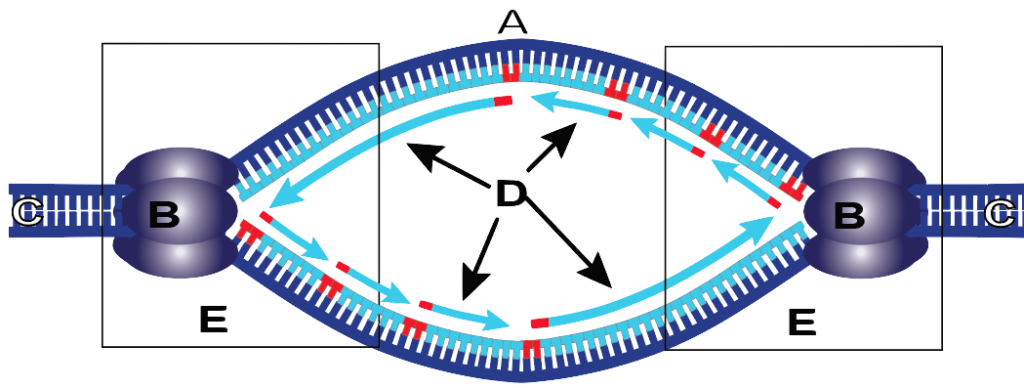
3. Replication forks are always created in pairs, creating a replication bubble (the entire structure below). In the replication bubble below, the original parent DNA is at C, helicase is at B, and newly synthesized DNA is shown in light blue at “D.” The two replication forks are at “E,” and the origin of replication is at “A.” In case you were wondering, we’ll get to the red nucleotides and the direction of the arrows in just a bit.
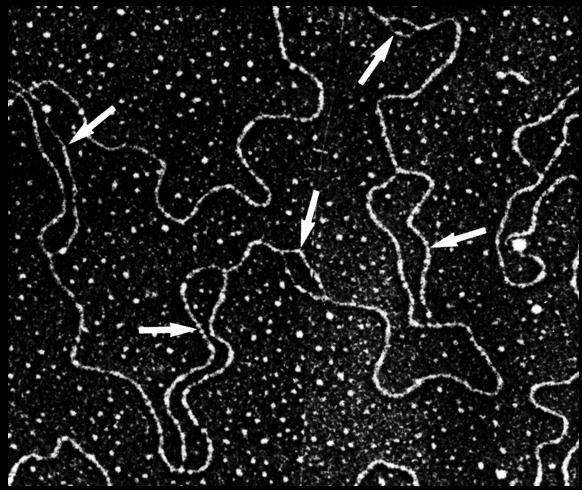
Check out this amazing electron micrograph. Each arrow is pointing to a replication bubble.
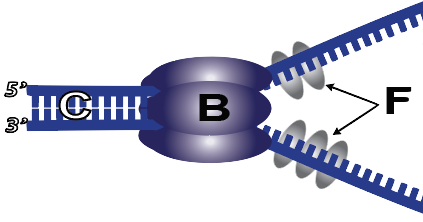
4. To keep the double helix from rewinding, two mechanisms are used. First, single-strand binding proteins (F) bind with the parent strand just behind helicase, preventing hydrogen bonds between opposite nucleotides from re-forming. In addition, an enzyme called topoisomerase (not shown) makes double-stranded or single-stranded cuts in DNA, allowing it to unwind so that other enzymes can assess the newly exposed single strands and replicate them. At the end of the process, other enzymes reseal the nick.
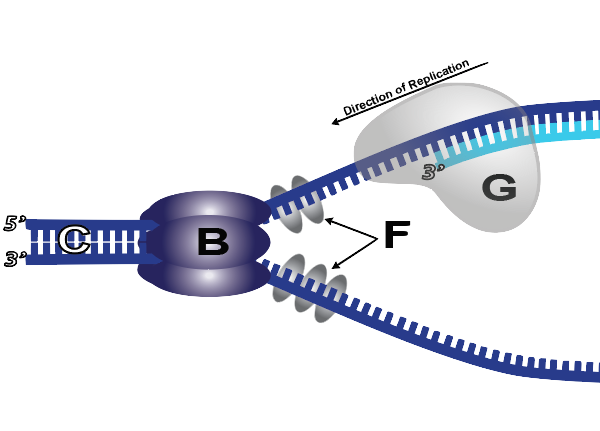
5. With the nitrogenous bases of the template (parent) strand exposed, the cell can now create complementary DNA on the daughter strand. The enzyme that does this is DNA polymerase III (“G” below). DNA polymerase III rides along the parent strand. As new nucleotides form hydrogen bonds with their complements on the template strand, DNA polymerase III catalyzes a sugar-phosphate bond between the growing strand and the new nucleotide at the 3′ end of the growing strand.
When thinking about this, it’s useful to imagine what DNA polymerase “knows” and “doesn’t know.” DNA polymerase doesn’t place the right nucleotide in position. Rather, nucleotides are floating around in the nucleus (or cytoplasm, in a prokaryotic cell). If the right nucleotide bumps into DNA at the right spot in the right orientation, it’ll temporarily hold in place with a hydrogen bond. At that moment, DNA polymerase’s active site will be correctly filled. That will cause DNA polymerase’s active site to change shape (by induced fit) and catalyze a sugar-phosphate bond between the new nucleotide and the growing strand.
6. DNA polymerase III can attach new nucleotides to a growing strand, but it can’t add a new nucleotide if there’s nothing to attach to. In other words, DNA polymerase’s active site requires that there be an existing strand behind it in the 5′ direction, and the correct new nucleotide ahead of it (in the 3′ direction). As a result, DNA polymerase III can’t initiate replication. Instead, another enzyme called primase (I) adds a short RNA primer (H) consisting of several RNA nucleotides. This primer holds onto the template strand through hydrogen bonds. Once the primer is in place, DNA polymerase III can start the replication process.
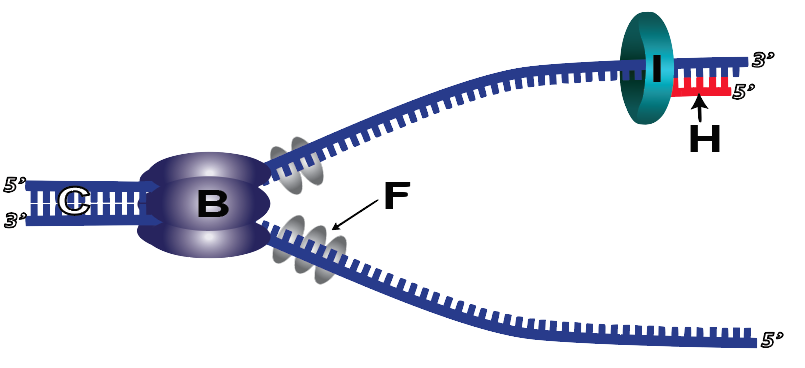
8. Because DNA polymerase III can only add new nucleotides at the 3′ end, a few more complications occur during DNA replication. The first has to do with what’s called the leading and lagging strands.
In the diagram below you see a replication bubble. The two DNA polymerase III enzymes that are following the opening replication forks are synthesizing new complementary DNA strands and attaching new nucleotides at the 3′ end. I’ve marked this new DNA with the letter “J.” This strand is known as the leading strand.
But how can DNA polymerase III synthesize a complementary DNA strand where I’ve put a question mark? On that side of the replication bubble, DNA polymerase III can’t follow helicase, because that would require DNA polymerase III to add new nucleotides at the 5′ end of a new strand which it can’t do.
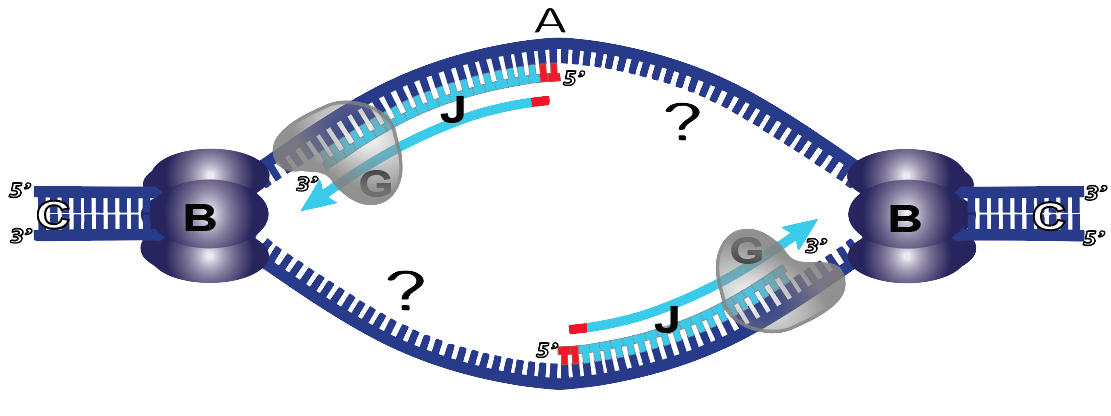
The strand where I’ve put a question mark is known as the lagging strand. It’s where synthesis moves away from the opening replication fork. This is in contrast to J, where DNA synthesis follows the replication fork.
The solution for the lagging strand is for DNA synthesis to proceed in fragments, shown below at “K.”
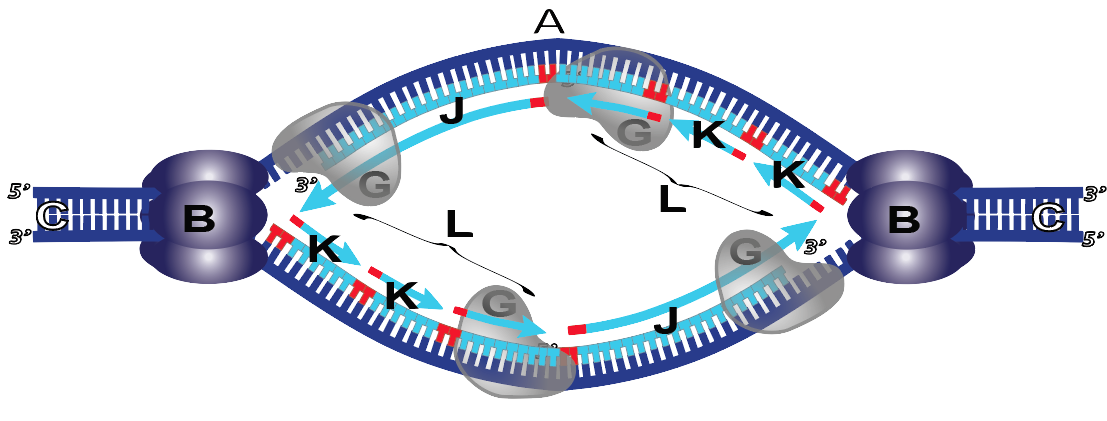
What you can see above is that on the leading strand (J), DNA synthesis is continuous. By contrast, in the lagging strand (L), DNA synthesis is fragmentary. Try to imagine the following: as the replication fork opens, Primase lays down its RNA primer. Then DNA polymerase comes in and lays down a short fragment of new DNA. When it reaches the RNA primer from another fragment, the DNA polymerase stops. So, fragment by fragment, DNA gets synthesized on the lagging strand. Note that in honor of their discoverer, Reiji Okazaki, these fragments are called “Okazaki fragments.”
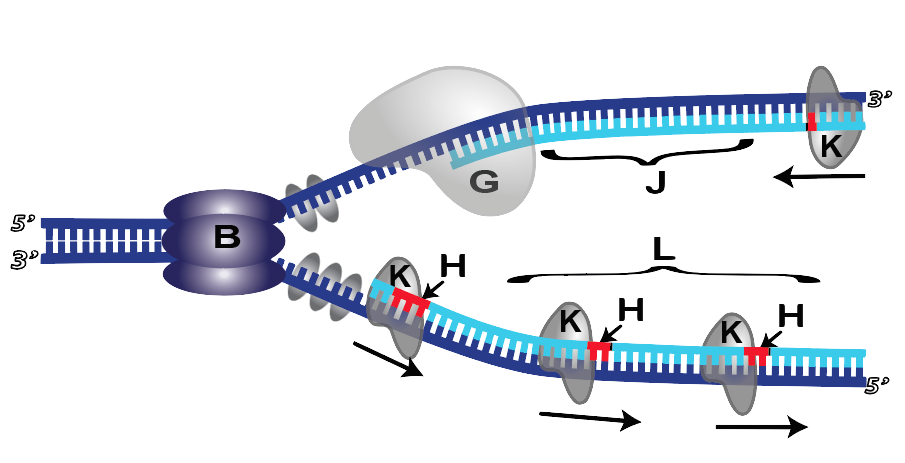
9. Because of Okazaki fragments and primers, there’s a lot of cleanup required at the end of the process. Cleanup begins as an enzyme called DNA polymerase I (shown at K below) comes and removes the primer, replacing RNA nucleotides with DNA nucleotides.
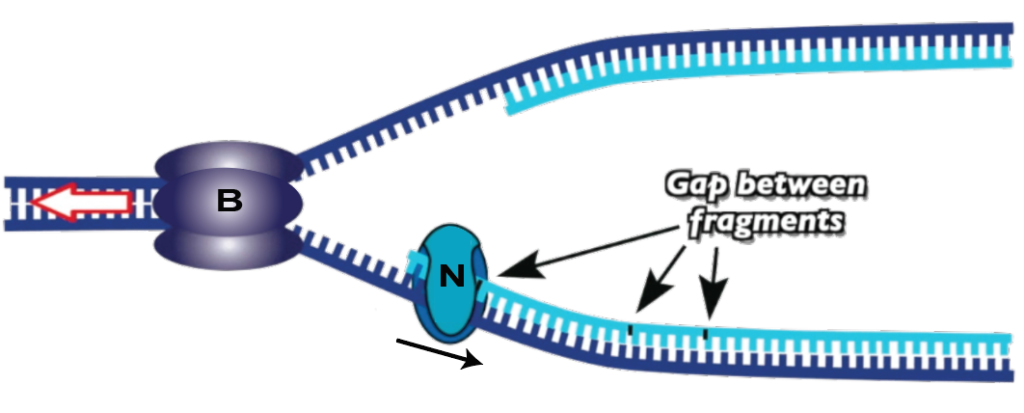
10. But even after the primer is removed, there’s still a gap (because DNA polymerase 1 can only add nucleotides to an existing strand: it can’t connect two strands together). To do this, a final enzyme, DNA ligase (N), comes and closes the gap between fragments.
4. A More Accurate Model
The process I described above is a great model for understanding DNA replication. But in cells, things are actually much more complex. Watch this computer animation from the Howard Hughes Medical Institute.
5. DNA Replication Music Video
Almost everything above, explained to a salsa beat. Click to read the lyrics.
6. Quiz: DNA Replication in Cells
The quiz below uses a combination of diagrams, multiple choice, and fill-in-the-blanks to help you consolidate your knowledge of DNA replication, as well as DNA structure.
[qwiz random=”true” style=”width: 550px !important; min-height: 400px !important;” qrecord_id=”sciencemusicvideosMeister1961-DNA Replication in Cells (v2.0)”]
[h]DNA Replication in Cells
[i]
[q]Which of the enzymes below is responsible for separating DNA into single strands so that each strand can be replicated?
[c]RE5BIHBvbHltZXJhc2UgSQ==[Qq]
[c]RE5BIHBvbHltZXJhc2UgSUlJ[Qq]
[c]aGVsaW Nhc2U=[Qq]
[c]bGlnYXNl[Qq]
[c]cHJpbWFzZQ==[Qq]
[c]c2luZ2xlLXN0cmFuZCBiaW5kaW5nIHByb3RlaW5z[Qq]
[f]Tm8uIEROQSBwb2x5bWVyYXNlIDEgaXMgdGhlIGVuenltZSByZXNwb25zaWJsZSBmb3IgcmVwbGFjaW5nIHRoZSBSTkEgcHJpbWVyIHdpdGggRE5BIG51Y2xlb3RpZGVzLiBMb29rIGF0IHRoZSBuYW1lcyBvZiB0aGVzZSBlbnp5bWVzIGFuZCB0aGluayAmIzgyMjA7d2hpY2ggb25lIA==YnJlYWtzIHRoZSBoZWxpeD8mIzgyMjE7[Qq]
[f]Tm8uIFBvbHltZXJhc2UgSUlJIGlzIHRoZSBtYWluIGVuenltZSByZXNwb25zaWJsZSBmb3IgYWRkaW5nIG5ldyBudWNsZW90aWRlcyBhdCB0aGUgMyYjODI0MjsgZW5kIG9mIGEgZ3Jvd2luZyBzdHJhbmQuIExvb2sgYXQgdGhlIG5hbWVzIG9mIHRoZXNlIGVuenltZXMgYW5kIHRoaW5rICYjODIyMDt3aGljaCBvbmUgYnJlYWtzIHRoZSBoZWxpeD8mIzgyMjE7[Qq]
[f]WWVzISBBcyBpdHMgbmFtZSBpbmRpY2F0ZXMsIGhlbGljYXNlIGlzIHRoZSBlbnp5bWUgdGhhdCBicmVha3MgdGhlIGhlbGl4Lg==[Qq]
[f]Tm8uIExpZ2FzZSBpcyB0aGUgZW56eW1lIHRoYXQgY29ubmVjdHMgb25lIEROQSBzZWN0aW9uIG9yIGZyYWdtZW50IHRvIGFub3RoZXIgb25lLiBMb29rIGF0IHRoZSBuYW1lcyBvZiB0aGVzZSBlbnp5bWVzIGFuZCB0aGluayAmIzgyMjA7d2hpY2ggb25lIA==YnJlYWtzIHRoZSBoZWxpeD8mIzgyMjE7[Qq]
[f]Tm8uIFByaW1hc2UgaXMgdGhlIGVuenltZSB0aGF0IGxheXMgZG93biBhIHByaW1lciBvZiBSTkEsIHNldHRpbmcgdGhlIHN0YWdlIGZvciBETkEgcG9seW1lcmFzZSBJSUkgdG8gYmVnaW4gc3RyYW5kIGVsb25nYXRpb24uIExvb2sgYXQgdGhlIG5hbWVzIG9mIHRoZXNlIGVuenltZXMgYW5kIHRoaW5rICYjODIyMDt3aGljaCBvbmUgYnJlYWtzIHRoZSBoZWxpeD8mIzgyMjE7[Qq]
[f]Tm8uIFNpbmdsZS1zdHJhbmQgYmluZGluZyBwcm90ZWlucyBiaW5kIHRvIHRoZSBleHBvc2VkIEROQSBzdHJhbmRzIGFuZCBwcmV2ZW50IHRoZW0gZnJvbSByZWJpbmRpbmcgdG8gb25lIGFub3RoZXIuIExvb2sgYXQgdGhlIG5hbWVzIG9mIHRoZXNlIGVuenltZXMgYW5kIHRoaW5rICYjODIyMDt3aGljaCBvbmUgYnJlYWtzIHRoZSBoZWxpeD8mIzgyMjE7[Qq]
[q]Which of the enzymes below is responsible for replacing the RNA primer with DNA nucleotides?
[c]RE5BIHBvbHlt ZXJhc2UgSQ==[Qq]
[c]RE5BIHBvbHltZXJhc2UgSUlJ[Qq]
[c]aGVsaWNhc2U=[Qq]
[c]bGlnYXNl[Qq]
[c]cHJpbWFzZQ==[Qq]
[c]c2luZ2xlIHN0cmFuZCBiaW5kaW5nIHByb3RlaW5z[Qq]
[f]WWVzLiBETkEgcG9seW1lcmFzZSAxIGlzIHRoZSBlbnp5bWUgcmVzcG9uc2libGUgZm9yIHJlcGxhY2luZyB0aGUgUk5BIHByaW1lciB3aXRoIEROQSBudWNsZW90aWRlcy4=[Qq]
[f]Tm8sIGJ1dCB5b3Ugd2VyZSBjbG9zZS4gRE5BIFBvbHltZXJhc2UgSUlJIGlzIHRoZSBtYWluIGVuenltZSByZXNwb25zaWJsZSBmb3IgYWRkaW5nIGFuZCBhdHRhY2hpbmcgbmV3IG51Y2xlb3RpZGVzIGF0IHRoZSAzJiM4MjQyOyBlbmQgb2YgYSBncm93aW5nIHN0cmFuZC4gTG9vayBhdCB0aGUgbmFtZXMgb2YgdGhlc2UgZW56eW1lcyBhbmQgYXNrIHlvdXJzZWxmICYjODIyMDt3aGljaCBvbmUgaXMgYWxzbyBsYXlpbmcgZG93biBuZXcgRE5BIG51Y2xlb3RpZGVzPyYjODIyMTs=[Qq]
[f]Tm8uIEFzIGl0cyBuYW1lIGluZGljYXRlcywgaGVsaWNhc2UgaXMgdGhlIGVuenltZSB0aGF0IGJyZWFrcyB0aGUgaGVsaXgu[Qq]
[f]Tm8uIExpZ2FzZSBpcyB0aGUgZW56eW1lIHRoYXQgY29ubmVjdHMgb25lIEROQSBzZWN0aW9uIG9yIGZyYWdtZW50IHRvIGFub3RoZXIgb25lLg==[Qq]
[f]Tm8uIFByaW1hc2UgaXMgdGhlIGVuenltZSB0aGF0IGxheXMgZG93biBhIHByaW1lciBvZiBSTkEsIHNldHRpbmcgdGhlIHN0YWdlIGZvciBETkEgcG9seW1lcmFzZSBJSUkgdG8gYmVnaW4gc3RyYW5kIGVsb25nYXRpb24u[Qq]
[f]Tm8uIFNpbmdsZS1zdHJhbmQgYmluZGluZyBwcm90ZWlucyBiaW5kIHRvIHRoZSBleHBvc2VkIEROQSBzdHJhbmRzIGFuZCBwcmV2ZW50IHRoZW0gZnJvbSByZWJpbmRpbmcgdG8gb25lIGFub3RoZXIu[Qq]
[q]Which of the enzymes below is the one that creates sugar-phosphate bonds connecting adjacent DNA fragments?
[c]RE5BIHBvbHltZXJhc2UgSQ==[Qq]
[c]RE5BIHBvbHltZXJhc2UgSUlJ[Qq]
[c]aGVsaWNhc2U=[Qq]
[c]bGln YXNl[Qq]
[c]cHJpbWFzZQ==[Qq]
[c]c2luZ2xlLXN0cmFuZCBiaW5kaW5nIHByb3RlaW5z[Qq]
[f]Tm8uIEROQSBwb2x5bWVyYXNlIDEgaXMgdGhlIGVuenltZSByZXNwb25zaWJsZSBmb3IgcmVwbGFjaW5nIHRoZSBSTkEgcHJpbWVyIHdpdGggRE5BIG51Y2xlb3RpZGVzLg==[Qq]
[f]Tm8sIEROQSBQb2x5bWVyYXNlIElJSSBpcyB0aGUgbWFpbiBlbnp5bWUgcmVzcG9uc2libGUgZm9yIGFkZGluZyBhdHRhY2hpbmcgbmV3IG51Y2xlb3RpZGVzIGF0IHRoZSAzJiM4MjQyOyBlbmQgb2YgYSBncm93aW5nIHN0cmFuZC4=[Qq]
[f]Tm8uIEFzIGl0cyBuYW1lIGluZGljYXRlcywgaGVsaWNhc2UgaXMgdGhlIGVuenltZSB0aGF0IGJyZWFrcyB0aGUgaGVsaXgu[Qq]
[f]WWVzLiBMaWdhc2UgaXMgdGhlIGVuenltZSB0aGF0IGNvbm5lY3RzIG9uZSBETkEgc2VjdGlvbiBvciBmcmFnbWVudCB0byBhbm90aGVyIG9uZS4=[Qq]
[f]Tm8uIFByaW1hc2UgaXMgdGhlIGVuenltZSB0aGF0IGxheXMgZG93biBhIHByaW1lciBvZiBSTkEsIHNldHRpbmcgdGhlIHN0YWdlIGZvciBETkEgcG9seW1lcmFzZSBJSUkgdG8gYmVnaW4gc3RyYW5kIGVsb25nYXRpb24u[Qq]
[f]Tm8uIFNpbmdsZS1zdHJhbmQgYmluZGluZyBwcm90ZWlucyBiaW5kIHRvIHRoZSBleHBvc2VkIEROQSBzdHJhbmRzIGFuZCBwcmV2ZW50IHRoZW0gZnJvbSByZWJpbmRpbmcgdG8gb25lIGFub3RoZXIu[Qq]
[q]Which of the enzymes below is the one that lays down a short primer of RNA, allowing DNA polymerase III to start attaching new DNA nucleotides to the growing strand?
[c]RE5BIHBvbHltZXJhc2UgSQ==[Qq]
[c]RE5BIHBvbHltZXJhc2UgSUlJ[Qq]
[c]aGVsaWNhc2U=[Qq]
[c]bGlnYXNl[Qq]
[c]cHJpbW FzZQ==[Qq]
[c]c2luZ2xlLXN0cmFuZCBiaW5kaW5nIHByb3RlaW5z[Qq]
[f]Tm8uIEROQSBwb2x5bWVyYXNlIDEgaXMgdGhlIGVuenltZSByZXNwb25zaWJsZSBmb3IgcmVwbGFjaW5nIHRoZSBSTkEgcHJpbWVyIHdpdGggRE5BIG51Y2xlb3RpZGVzLiBGb3IgdGhpcyBlbnp5bWUsIGp1c3QgZmluZCBhbiBlbnp5bWUgd2hvc2UgbmFtZSBpcyBzaW1pbGFyIHRvICYjODIyMDtwcmltZXImIzgyMjE7IGFuZCB5b3UmIzgyMTc7bGwgaGF2ZSB5b3VyIGFuc3dlci4=[Qq]
[f]Tm8sIEROQSBQb2x5bWVyYXNlIElJSSBpcyB0aGUgbWFpbiBlbnp5bWUgcmVzcG9uc2libGUgZm9yIGFkZGluZyBhdHRhY2hpbmcgbmV3IG51Y2xlb3RpZGVzIGF0IHRoZSAzJiM4MjQyOyBlbmQgb2YgYSBncm93aW5nIHN0cmFuZC4gRm9yIHRoaXMgZW56eW1lLCBqdXN0IGZpbmQgYW4gZW56eW1lIHdob3NlIG5hbWUgaXMgc2ltaWxhciB0byAmIzgyMjA7cHJpbWVyJiM4MjIxOyBhbmQgeW91JiM4MjE3O2xsIGhhdmUgeW91ciBhbnN3ZXIu[Qq]
[f]Tm8uIEFzIGl0cyBuYW1lIGluZGljYXRlcywgaGVsaWNhc2UgaXMgdGhlIGVuenltZSB0aGF0IGJyZWFrcyB0aGUgaGVsaXguIEZvciB0aGlzIGVuenltZSwganVzdCBmaW5kIGFuIGVuenltZSB3aG9zZSBuYW1lIGlzIHNpbWlsYXIgdG8gJiM4MjIwO3ByaW1lciYjODIyMTsgYW5kIHlvdSYjODIxNztsbCBoYXZlIHlvdXIgYW5zd2VyLg==[Qq]
[f]Tm8uIExpZ2FzZSBpcyB0aGUgZW56eW1lIHRoYXQgY29ubmVjdHMgb25lIEROQSBzZWN0aW9uIG9yIGZyYWdtZW50IHRvIGFub3RoZXIgb25lLiBGb3IgdGhpcyBlbnp5bWUsIGp1c3QgZmluZCBhbiBlbnp5bWUgd2hvc2UgbmFtZSBpcyBzaW1pbGFyIHRvICYjODIyMDtwcmltZXImIzgyMjE7IGFuZCB5b3UmIzgyMTc7bGwgaGF2ZSB5b3VyIGFuc3dlci4=[Qq]
[f]RXhjZWxsZW50LiBQcmltYXNlIGlzIHRoZSBlbnp5bWUgdGhhdCBsYXlzIGRvd24gYSBwcmltZXIgb2YgUk5BLCBzZXR0aW5nIHRoZSBzdGFnZSBmb3IgRE5BIHBvbHltZXJhc2UgSUlJIHRvIGJlZ2luIHN0cmFuZCBlbG9uZ2F0aW9uLg==[Qq]
[f]Tm8uIFNpbmdsZS1zdHJhbmQgYmluZGluZyBwcm90ZWlucyBhcmUgZW56eW1lcyB0aGF0IGJpbmQgdG8gdGhlIGV4cG9zZWQgRE5BIHN0cmFuZHMgYW5kIHByZXZlbnQgdGhlbSBmcm9tIHJlYmluZGluZyB0byBvbmUgYW5vdGhlci4gRm9yIHRoaXMgZW56eW1lLCBqdXN0IGZpbmQgYW4gZW56eW1lIHdob3NlIG5hbWUgaXMgc2ltaWxhciB0byAmIzgyMjA7cHJpbWVyJiM4MjIxOyBhbmQgeW91JiM4MjE3O2xsIGhhdmUgeW91ciBhbnN3ZXIu[Qq]
[q]Which of the molecules below bind to the exposed DNA strands and prevent them from rebinding to one another?
[c]RE5BIHBvbHltZXJhc2UgSQ==[Qq]
[c]RE5BIHBvbHltZXJhc2UgSUlJ[Qq]
[c]aGVsaWNhc2U=[Qq]
[c]bGlnYXNl[Qq]
[c]cHJpbWFzZQ==[Qq]
[c]c2luZ2xlLXN0cmFuZCBi aW5kaW5nIHByb3RlaW5z[Qq]
[f]Tm8uIEROQSBwb2x5bWVyYXNlIDEgaXMgdGhlIGVuenltZSByZXNwb25zaWJsZSBmb3IgcmVwbGFjaW5nIHRoZSBSTkEgcHJpbWVyIHdpdGggRE5BIG51Y2xlb3RpZGVzLg==[Qq]
[f]Tm8sIEROQSBQb2x5bWVyYXNlIElJSSBpcyB0aGUgbWFpbiBlbnp5bWUgcmVzcG9uc2libGUgZm9yIGFkZGluZyBhdHRhY2hpbmcgbmV3IG51Y2xlb3RpZGVzIGF0IHRoZSAzJiM4MjQyOyBlbmQgb2YgYSBncm93aW5nIHN0cmFuZC4=[Qq]
[f]Tm8uIEFzIGl0cyBuYW1lIGluZGljYXRlcywgaGVsaWNhc2UgaXMgdGhlIGVuenltZSB0aGF0IGJyZWFrcyB0aGUgaGVsaXgu[Qq]
[f]Tm8uIExpZ2FzZSBpcyB0aGUgZW56eW1lIHRoYXQgY29ubmVjdHMgb25lIEROQSBzZWN0aW9uIG9yIGZyYWdtZW50IHRvIGFub3RoZXIgb25lLg==[Qq]
[f]Tm8uIFByaW1hc2UgaXMgdGhlIGVuenltZSB0aGF0IGxheXMgZG93biBhIHByaW1lciBvZiBSTkEsIHNldHRpbmcgdGhlIHN0YWdlIGZvciBETkEgcG9seW1lcmFzZSBJSUkgdG8gYmVnaW4gc3RyYW5kIGVsb25nYXRpb24u[Qq]
[f]WWVzLiBTaW5nbGUtc3RyYW5kIGJpbmRpbmcgcHJvdGVpbnMgYXJlIGVuenltZXMgdGhhdCBiaW5kIHRvIHRoZSBleHBvc2VkIEROQSBzdHJhbmRzIGFuZCBwcmV2ZW50IHRoZW0gZnJvbSByZWJpbmRpbmcgdG8gb25lIGFub3RoZXIu
Cg==[Qq]
[q]In the diagram below, which letter is helicase?
[textentry single_char=”true”]
[c]Qg ==[Qq]
[f]IFllcy4g4oCcQuKAnSBpcyBoZWxpY2FzZS4=[Qq]
[c]Kg==[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIEhlbGljYXNlIGlzIHRoZSBlbnp5bWUgdGhhdCBvcGVucyB1cCB0aGUgaGVsaXguIE9mIHRoZSBsZXR0ZXJlZCBpdGVtcyBpbiB0aGlzIGRpYWdyYW0sIHdoaWNoIGlzIHRoZSBvbmx5IG9uZSB0aGF0IGNvdWxkIGJlIGRvaW5nIHRoYXQ/[Qq]
[q]In the diagram below, which letter is showing the origin of replication?
[textentry single_char=”true”]
[c]QQ ==[Qq]
[f]IFllcy4g4oCcQeKAnSBpcyB0aGUgb3JpZ2luIG9mIHJlcGxpY2F0aW9uLg==[Qq]
[c]Kg==[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIExvb2sgYXQgdGhlIGRpYWdyYW0gYmVsb3cuIFRoZSBvcmlnaW4gaXMgd2hlcmUgcmVwbGljYXRpb24gc3RhcnRzLiBOZXh0IHRpbWUgeW91IHNlZSB0aGlzIHF1ZXN0aW9uLCBhcHBseSB3aGF0JiM4MjE3O3Mgb2J2aW91c2x5IHRoZSBvcmlnaW4gaW4gdGhlIGRpYWdyYW0gYmVsb3cgdG8gdGhlIGRpYWdyYW0gYWJvdmUu[Qq]
[q]In the diagram below, which letter indicates a replication fork?
[textentry single_char=”true”]
[c]RQ ==[Qq]
[f]IFllcy4g4oCcReKAnSBpcyBhIHJlcGxpY2F0aW9uIGZvcmsu[Qq]
[c]Kg==[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIFRoZSByZXBsaWNhdGlvbiBmb3JrIGlzIHdoZXJlIHRoZSBzaW5nbGUgRE5BIHN0cmFuZCBpcyBzcGxpdCBvcGVuIGludG8gdHdvIHRlbXBsYXRlIHN0cmFuZHMuIFdpdGggdGhhdCBpbiBtaW5kLCB3aGF0IGFyZSB0aGUgb25seSBsZXR0ZXJzIGluIHRoaXMgZGlhZ3JhbSB0aGF0IGNvdWxkIHJlcHJlc2VudCBzb21ldGhpbmcgdGhhdCYjODIxNztzIGxpa2UgYSBmb3JrPw==[Qq]
[q]In the diagram below, which number indicates a replication fork?
[textentry single_char=”true”]
[c]Nw ==[Qq]
[f]IFllcy4g4oCcN+KAnSBpcyBhIHJlcGxpY2F0aW9uIGZvcmsu[Qq]
[c]Kg==[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIFRoZSByZXBsaWNhdGlvbiBmb3JrIGlzIHdoZXJlIHRoZSBzaW5nbGUgRE5BIHN0cmFuZCBpcyBzcGxpdCBvcGVuIGludG8gdHdvIHRlbXBsYXRlIHN0cmFuZHMuIFdpdGggdGhhdCBpbiBtaW5kLCB3aGF0IGlzIHRoZSBvbmx5IG51bWJlciBpbiB0aGlzIGRpYWdyYW0gdGhhdCBjb3VsZCByZXByZXNlbnQgc29tZXRoaW5nIHRoYXQmIzgyMTc7cyBsaWtlIGEgZm9yaz8=[Qq]
[q]In the diagram below, which number indicates helicase?
[textentry single_char=”true”]
[c]MQ ==[Qq]
[f]IFllcy4g4oCcMeKAnSBpcyBoZWxpY2FzZQ==[Qq]
[c]Kg==[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIEhlbGljYXNlIGlzIHRoZSBlbnp5bWUgdGhhdCBzcGxpdHMgb3BlbiB0aGUgcGFyZW50YWwgRE5BLCBjcmVhdGluZyB0aGUgcmVwbGljYXRpb24gZm9yay4=[Qq]
[q]In the diagram below, which number is DNA Polymerase III?
[textentry single_char=”true”]
[c]Mg ==[Qq]
[f]IFllcy4g4oCcMuKAnSBpcyBETkEgcG9seW1lcmFzZSBJSUksIHdoaWNoIHN5bnRoZXNpemVzIG5ldyBETkEgYnkgYXR0YWNoaW5nIG5ldyBudWNsZW90aWRlcyB0byB0aGUgMyYjODI0MjsgZW5kIG9mIGEgZ3Jvd2luZyBETkEgc3RyYW5kLg==[Qq]
[c]Kg==[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIEROQSBwb2x5bWVyYXNlIHN5bnRoZXNpemVzIG5ldyBETkEgYnkgYXR0YWNoaW5nIG5ldyBudWNsZW90aWRlcyB0byB0aGUgMyYjODI0MjsgZW5kIG9mIGEgRE5BIHN0cmFuZC4gVGhlIG5ldyBudWNsZW90aWRlcyBhcmUgc2hvd24gaW4gbGlnaHQgYmx1ZS4gV2hpY2ggZW56eW1lIHNob3duIGFib3ZlIHNlZW1zIHRvIGJlIGludm9sdmVkIHdpdGggdGhlc2UgbmV3IG51Y2xlb3RpZGVzPw==[Qq]
[q]In the diagram below, which number indicates the lagging strand?
[textentry single_char=”true”]
[c]Mw ==[Qq]
[f]IFllcy4g4oCcM+KAnSBpcyB0aGUgbGFnZ2luZyBzdHJhbmQu[Qq]
[c]Kg==[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIFRoZSBsYWdnaW5nIHN0cmFuZCBpcyB3aGVyZSBETkEgc3ludGhlc2lzIG9jY3VycyBpbiBhIGRpcmVjdGlvbiBvcHBvc2l0ZSB0byB0aGF0IG9mIHRoZSB0aGUgb3BlbmluZyByZXBsaWNhdGlvbiBmb3JrLCBhbmQsIGFzIGEgcmVzdWx0LCBoYXMgdG8gYmUgZnJhZ21lbnRhcnkuIFdoZXJlIGRvIHlvdSBzZWUgc3ludGhlc2lzIG1vdmluZyBhd2F5IGZyb20gdGhlIHJlcGxpY2F0aW9uIGZvcms/[Qq]
[q]In the diagram below, which number indicates an RNA primer?
[textentry single_char=”true”]
[c]NA ==[Qq]
[f]IFllcy4g4oCcNOKAnSBpcyBhbiBSTkEgcHJpbWVyLg==[Qq]
[c]Kg==[Qq]
[f]Tm8uIEhlcmUmIzgyMTc7cyBhIGhpbnQuIEROQSBwb2x5bWVyYXNlIElJSSBjYW4gb25seSBhdHRhY2ggbnVjbGVvdGlkZXMgdG8gYW4gZXhpc3Rpbmcgc3RyYW5kLiBJdCBjYW4mIzgyMTc7dCBpbml0aWF0ZSBhIG5ldyBzdHJhbmQgd2hlcmUgbm9uZSBleGlzdHMuIEluc3RlYWQsIGFuIGVuenltZSBjYWxsZWQgcHJpbWFzZSAoNSkgbGF5cyBkb3duIGEgc2hvcnQgc3RyYW5kIG9mIFJOQSAodGhlIHByaW1lciksIHdoaWNoIEROQSBwb2x5bWVyYXNlIElJSSB3aWxsIHVzZSBhcyBhIHN0YXJ0aW5nIHBvaW50IGZvciByZXBsaWNhdGlvbi4gRmluZCAjIDUgYW5kIHlvdSYjODIxNztsbCBoYXZlIHlvdXIgYW5zd2VyLg==[Qq]
[q]In the diagram below, which number indicates the leading strand?
[textentry single_char=”true”]
[c]Ng ==[Qq]
[f]IFllcy4g4oCcNuKAnSBpcyB0aGUgbGVhZGluZyBzdHJhbmQu[Qq]
[c]Kg==[Qq]
[f]Tm8uIFRoZSBsZWFkaW5nIHN0cmFuZCBpcyB0aGUgc3RyYW5kIHdoZXJlIEROQSBwb2x5bWVyYXNlIGlzIGZvbGxvd2luZyBoZWxpY2FzZSBhdCB0aGUgdGhlIG9wZW5pbmcgcmVwbGljYXRpb24gZm9yay4gRmluZCB0aGUgRE5BIHBvbHltZXJhc2UgdGhhdCYjODIxNztzIGZvbGxvd2luZyB0aGUgZm9yaywgYW5kIHlvdSYjODIxNztsbCBoYXZlIHlvdXIgYW5zd2VyLg==[Qq]
[q]In the diagram below, which number is pointing to the single-strand binding proteins?
[textentry single_char=”true”]
[c]OA ==[Qq]
[f]IFllcy4g4oCcOOKAnSBpcyBwb2ludGluZyB0byB0aGUgc2luZ2xlLXN0cmFuZCBiaW5kaW5nIHByb3RlaW5z[Qq]
[c]Kg==[Qq]
[f]Tm8uIFRoZSBzaW5nbGUtc3RyYW5kIGJpbmRpbmcgcHJvdGVpbnMgYmluZCB0byB0aGUgRE5BIGF0IHRoZSByZXBsaWNhdGlvbiBmb3JrLCBrZWVwaW5nIHRoZSBETkEgZnJvbSBjbG9zaW5nIHVwICh3aGljaCB3b3VsZCBlbmQgdGhlIHJlcGxpY2F0aW9uIHByb2Nlc3MuIExvb2sgZm9yIHRoZXNlIHByb3RlaW5zIGNsb3NlIHRvIHRoZSBvcGVuaW5nIG9mIHRoZSBmb3JrLg==
[Qq]
[q]In the diagram of a replication bubble below, which letter is pointing to the leading strand?
[textentry single_char=”true”]
[c]Sg ==[Qq]
[f]IFllcy4g4oCcSuKAnSBpcyBwb2ludGluZyB0byB0aGUgbGVhZGluZyBzdHJhbmQuIFlvdSBjYW4gdGVsbCBiZWNhdXNlIHRoZSBzdHJhbmQgaXMgYmVpbmcgc3ludGhlc2l6ZWQgaW4gdGhlIHNhbWUgZGlyZWN0aW9uIGFzIHRoZSBvcGVuaW5nIHJlcGxpY2F0aW9uIGZvcmsu[Qq]
[c]Kg==[Qq]
[f]Tm8uIEZpbmQgdGhlIHN0YW5kIHdoZXJlIHN5bnRoZXNpcyBpcyBjb250aW51b3VzLCBhcyBETkEgcG9seW1lcmFzZSBmb2xsb3dzIGhlbGljYXNlLg==
Cg==[Qq]
[q]In the diagram of a replication bubble below, which letter is labeling an Okazaki fragment?
[textentry single_char=”true”]
[c]Sw ==[Qq]
[f]IFllcy4g4oCcS+KAnSBpcyBsYWJlbGluZyBhbiBPa2F6YWtpIGZyYWdtZW50LCBvbmUgb2YgdGhlIGZyYWdtZW50cyBvZiBETkEgc3ludGhlc2l6ZWQgYWxvbmcgdGhlIGxhZ2dpbmcgc3RyYW5kLg==[Qq]
[c]Kg==[Qq]
[f]Tm8uIEZpbmQgdGhlIHNob3J0IHN0cmV0Y2hlcyBvZiBmcmFnbWVudGFyeSBETkEgdGhhdCBhcmUgc3ludGhlc2l6ZWQgb24gdGhlIGxhZ2dpbmcgc3RyYW5kICh0aGUgc3RyYW5kIHdoZXJlIHN5bnRoZXNpcyBpcyBtb3ZpbmcgYXdheQ==IGZyb20gdGhlIHJlcGxpY2F0aW9uIGZvcmspLg==
Cg==[Qq]
[q]In the diagram of a replication bubble below, which letter is pointing to the lagging strand?
[textentry single_char=”true”]
[c]TA ==[Qq]
[f]IFllcy4g4oCcTOKAnSBpcyBwb2ludGluZyB0byB0aGUgbGFnZ2luZyBzdHJhbmQu[Qq]
[c]Kg==[Qq]
[f]Tm8uIEZpbmQgdGhlIHNob3J0IHN0cmV0Y2hlcyBvZiBmcmFnbWVudGFyeSBETkEuIFRoZXNlIGFyZSBPa2F6YWtpIGZyYWdtZW50cywgYW5kIHRoZXkmIzgyMTc7cmUgb25seSBzeW50aGVzaXplZCBvbiB0aGUgbGFnZ2luZyBzdHJhbmQu
Cg==
[q labels = “top”]
[l]DNA helicase
[fx] No. Please try again.
[f*] Good!
[l]parent strand
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]DNA polymerase III
[fx] No. Please try again.
[f*] Great!
[l]newly synthesized DNA
[fx] No. Please try again.
[f*] Excellent!
[q]Each of the following is a step in DNA replication. The correct order of these steps is
I. An enzyme connects the sugars and phosphates of adjacent nucleotides.
II. Following the base pairing rules, new nucleotides bond with the template strand
III. An enzyme separates the two strands
[c]SSwgSUksIElJSQ==[Qq]
[c]SUlJLCBJ SSwgSQ==[Qq]
[c]SUksIElJSSwgSQ==[Qq]
[f]Tm8uIFlvdSBoYXZlIHRoZSBvcmRlciByZXZlcnNlZC4=[Qq]
[f]WWVzLiBEdXJpbmcgcmVwbGljYXRpb24sIGZpcnN0IHRoZSBzdHJhbmRzIGFyZSBzZXBhcmF0ZWQuIFRoZW4gbmV3IG51Y2xlb3RpZGVzIGJvbmQuIFRoZW4gYW4gZW56eW1lIGNyZWF0ZXMgc3VnYXItcGhvc3BoYXRlIGJvbmRzIGJldHdlZW4gYWRqYWNlbnQgbnVjbGVvdGlkZXMu[Qq]
[f]Tm8uIEZvciBzdGVwIElJIHRvIG9jY3VyLCB0aGUgc3RyYW5kIG5lZWRzIHRvIGJlIG9wZW5lZCBzbyB0aGF0IG5ldyBudWNsZW90aWRlcyBjYW4gYm9uZC4=[Qq]
[q]The result of DNA replication is a new molecule composed of one new strand, and one old strand (from the parent molecule). A way to describe this is ______-______ replication.
[hangman]
[c]c2VtaS1jb25zZXJ2YXRpdmU=[Qq]
[f]R29vZCBKb2IhIFRoZSByZXN1bHQgb2YgRE5BIHJlcGxpY2F0aW9uIGlzIGEgbmV3IG1vbGVjdWxlIGNvbXBvc2VkIG9uIG9uZSBuZXcgc3RyYW5kLCBhbmQgb25lIG9sZCBzdHJhbmQgKGZyb20gdGhlIHBhcmVudCBtb2xlY3VsZSkuIEEgd2F5IHRvIGRlc2NyaWJlIHRoaXMgaXMgc2VtaS1jb25zZXJ2YXRpdmU=IHJlcGxpY2F0aW9u
Cg==[Qq]
[q]The bonds that helicase breaks are [hangman] bonds.
[c]aHlkcm9nZW4=[Qq]
[f]UGVyZmVjdCEgVGhlIGJvbmRzIHRoYXQgaGVsaWNhc2UgYnJlYWtzIGFyZSA=aHlkcm9nZW4=IGJvbmRzLg==[Qq]
[q]During replication, each strand serves as a [hangman] for the synthesis of a second strand.
[c]dGVtcGxhdGU=
Cg==[Qq]
[q]During DNA replication, the key enzymes always move in a [hangman] prime to [hangman] prime direction (hint: write out the numbers).
[c]Zml2ZQ==[Qq]
[c]dGhyZWU=
Cg==[Qq]
[q]When nucleotides are added to a growing DNA strand, they’re always added on the [hangman] prime side.(hint: write out the number).
[c]dGhyZWU=
Cg==[Qq]
[q]The enzyme that opens up the double helix is [hangman].
[c]aGVsaWNhc2U=[Qq]
[q]Helicase first opens up the double helix at a sequence of bases called the origin of [hangman].
[c]cmVwbGljYXRpb24=
Cg==Cg==[Qq]
[q]The structure formed by two replication forks is called a replication [hangman].
[c]YnViYmxl
Cg==[Qq]
[q]During replication, [hangman]-[hangman]-[hangman]-[hangman] prevent the double helix from rewinding.
[c]c2luZ2xl[Qq]
[c]c3RyYW5k[Qq]
[c]YmluZGluZw==[Qq]
[c]cHJvdGVpbnM=
Cg==[Qq]
[q]After helicase opens up the double helix, [hangman] lays down a primer of RNA.
[c]cHJpbWFzZQ==[Qq]
[q]The job of DNA [hangman] 1 is to remove the RNA primer.
[c]cG9seW1lcmFzZQ==[Qq]
[q]The enzyme whose job it is to connect the fragments created during replication is [hangman].
[c]bGlnYXNl[Qq]
[x][restart]
[/qwiz]
7. DNA Replication Flashcards
One of the flashcards below focuses on the Meselson-Stahl experiment, also called the Pulse-Chase Primer Experiment. In this experiment, performed in 1957, Matthew Meselson and Franklin Stahl proved that the semi-conservative model of DNA replication was correct.
To learn about this experiment, please watch this video by Paul Anderson. You can also consult this article on Wikipedia. Both links open up in a new browser tab.
[qdeck bold_text=”false” style=”width: 625px !important; min-height: 475px !important;” qrecord_id=”sciencemusicvideosMeister1961-DNA Replication Flashcards (v2.0)”]
[h]DNA Replication Flashcards
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|1925a55316110″ question_number=”211″ topic=”6.1-2.DNA_and_RNA,_DNA_Replication”] DNA replication is semiconservative. On a big-picture level, describe how semiconservative replication occurs (and what that term means).
[a] Durin DNA replication, a team of enzymes, using each strand of the double helix as a template, synthesizes new daughter strands. As a result, each daughter DNA double helix consists of one conserved strand from the parent molecule, and another strand that was synthesized anew, making the process semi-conservative.
[q]Describe the roles of DNA polymerase, helicase, and single-strand binding proteins in DNA replication.
[a]Helicase opens up helix at the origin of replication, creating a replication fork. DNA polymerase creates sugar-phosphate bonds between a new nucleotide at the 3′ end of a new strand, and an existing DNA strand, building a new daughter strand in the 5′ to 3′ direction. Single-strand binding proteins keep the double helix from rewinding as replication occurs.
[q]What is primase, and why is it needed?
[a]DNA polymerase III can attach new nucleotides to a growing strand, but it can’t add a new nucleotide if there’s nothing to attach to. In other words, DNA polymerase’s active site requires that there be an existing strand behind it in the 5′ direction, and the correct new nucleotide ahead of it (in the 3′ direction). As a result, DNA polymerase III can’t initiate replication. Instead, another enzyme called primase (I) adds a short RNA primer (H) consisting of several RNA nucleotides. This primer holds onto the template strand through hydrogen bonds. Once the primer is in place, DNA polymerase III can start the replication process.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|191c9fa502910″ question_number=”212″ topic=”6.1-2.DNA_and_RNA,_DNA_Replication”] Describe how DNA replication starts. End with the binding of DNA polymerase. In your response, include the roles of the following: the origin of replication, replication fork, RNA primase, and primer.
[a] Replication begins when an enzyme called helicase (1) finds a sequence called the origin of replication and separates the double-stranded DNA, exposing two single strands in a structure called a replication fork (7). An enzyme called RNA primase (5) lays down a short stretch of complementary RNA called a primer (4). The primer enables DNA polymerase (2) to bind and to use the template strand (dark blue) as a guide for synthesizing a complementary strand (light blue), always adding new nucleotides at the 3’ end of a growing strand.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|191374b630d10″ question_number=”213″ topic=”6.1-2.DNA_and_RNA,_DNA_Replication”] How is DNA replication at the leading strand different from replication at the lagging strand?
[a] In the leading strand (4), DNA replication is relatively continuous, as DNA polymerase (3) follows the opening replication fork. In the lagging strand, DNA polymerase synthesizes in the opposite direction from the opening replication fork. This results in short sequences that are called Okazaki fragments (5).
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|190a49c75f110″ question_number=”214″ topic=”6.1-2.DNA_and_RNA,_DNA_Replication”] Describe the roles of ligase and topoisomerase in DNA replication.
[a] After DNA polymerase has laid down all of the complementary nucleotides that it can, an enzyme called ligase (N) creates a sugar-phosphate bond between DNA fragments. Throughout the replication process, enzymes called topoisomerases (not shown) nick DNA’s sugar-phosphate backbone, preventing the DNA from overwinding, and then resealing the break.
[q] The diagram below depicts semi-conservative replication of DNA.
Explain how the Meselson-Stahl experiment proved the semiconservative model to be correct.
[a] Meselson and Stahl started by growing a culture of E. coli on N15, a heavy isotope of nitrogen. As a result, all of the nitrogen in this population of E. coli’s DNA was composed of this heavier isotope. Then they transferred this E. coli to a medium containing N14 (a lighter isotope). They allowed the E. coli to replicate itself once and then used centrifugation to determine the weight of the nitrogen in the E. coli’s DNA. They found that the DNA’s weight was halfway between N14 and N15. This established that the new DNA was composed of 50% N14 and 50%, N15, proving the conservative model of DNA replication was incorrect. Why? Because conservative replication would result in some DNA that was all composed of N15 (DNA that was conserved), with other DNA being composed exclusively of N14 (the newly synthesized DNA).
Meselson and Stahl then let the bacteria replicate for a second generation and again sampled the DNA. They found that in the second generation, there continued to be DNA whose weight was halfway between N14 and N15, and new DNA that was completely N14. This proved the dispersive model to be incorrect and corroborated what one would predict if DNA were semi-conservatively replicated.
[x][restart]
[/qdeck]
What’s next?
Proceed to Topics 6.3 and 6.4, part 1: Transcription (the next tutorial in AP Bio Unit 6).