1. Introduction
In previous tutorials in AP Bio Topics 6.5 and 6.6, we’ve seen how gene expression in eukaryotes involves
- epigenetic modifications to DNA, plus
- a variety of mechanisms involving transcription factors that allow eukaryotes to control which genes in a particular type of tissue are transcribed.
In what’s below, you’ll learn about two more mechanisms for the regulation of eukaryotic gene expression.
2. Coordinated Control of Gene Expression
Throughout the life of a eukaryotic organism, there are many occasions where different genes need to be turned on and off together. These genes may be in the same cell but on different chromosomes. Or they may be in different tissues in widely separated parts of the body. How can this coordination occur?
Unlike prokaryotic cells, these genes are not clustered together in operons. The mechanism for turning these separate genes on at once involves shared regulatory sequences that respond to the same transcription factors
2a. Coordinated control of gene expression in plants: the drought response element

One example of coordinated gene expression involves how plants respond to stress induced by drought. To survive, plants must produce a variety of proteins simultaneously.
To do this, inactive transcription factors (shown at “a”, below) have to respond to stress by changing to an activated form (at “b”). This activation may involve phosphorylation or another type of conformational change.
Once activated, these transcription factors will bind with any promoter that has, as part of its structure, a matching regulatory nucleotide sequence. In the diagram above, you can see three genes, each of which has a promoter (d) containing what’s called a “dehydration response element” (indicated by “c”). The binding of the transcription factor with this element allows RNA polymerase (g) to bind, producing each gene’s RNA (at “e”). This mRNA then gets translated into a variety of proteins (at “f”).
2b. Coordinated Control of gene expression in animals: steroid hormones and gene activation
|
|
|
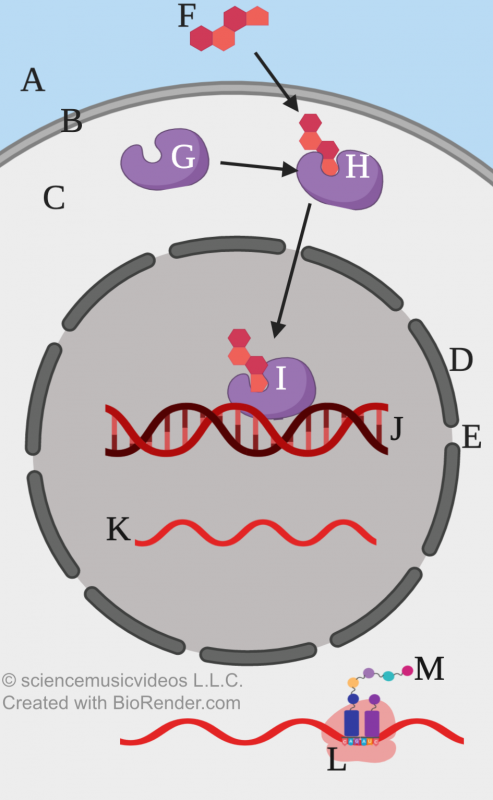
In our bodies (and the bodies of related animals), a similar mechanism explains how one hormone can activate a variety of genes in a variety of cells. An example is the steroid hormone estrogen. Estrogen is known as the feminizing hormone, and in females, it’s involved in the development of secondary sexual characteristics, ovulation, changes in the uterine lining during the menstrual cycle, and sexual receptivity.
Estrogen is produced primarily by ovaries (but also by the placenta during pregnancy). From the ovaries, it diffuses into the bloodstream and then goes everywhere in the body.
As a steroid hormone, estrogen can diffuse through the cell’s phospholipid bilayer. In the cytoplasm, estrogen binds with an estrogen receptor.
The estrogen receptor, once bound to estrogen, becomes a transcription factor. It diffuses into the cell’s nucleus, where it binds with estrogen response elements. These control elements combine with other transcription factors to create a transcription initiation complex, resulting in the transcription of estrogen-related genes.
The diagram on the left shows a generalized version of this process. “F” represents estrogen. “G” is the estrogen receptor. Once the receptor binds to estrogen, it becomes a transcription factor. Letter “H” shows this transcription factor: a cytoplasmic receptor bound to estrogen. This transcription factor can cross the nuclear membrane (“D”). “I” shows this transcription factor interacting with DNA (“J”), producing RNA that can (after RNA processing) be translated by ribosomes (“L”) into protein (“M”).
The mechanism by which the masculinizing hormone testosterone works is similar.
What’s the takeaway? Hormones can act as transcription factors, activating a variety of genes throughout the body to create coordinated responses to stimuli that can come from inside or outside the body. In the case of the steroid hormones estrogen and testosterone, these hormones cause changes throughout the body at several moments during development. These include initial sexual differentiation, which occurs, in humans, about six weeks after conception. The second round of sexual differentiation occurs during puberty when secondary sexual characteristics develop in response to another release of estrogen and testosterone.
3. Quiz: Coordinated control of transcription
[qwiz random = “true” style=”width: 650px !important; min-height: 400px !important;” qrecord_id=”sciencemusicvideosMeister1961-Coordinated Control of Eukaryotic Transcription (v2.0)”] [h]
Coordinated Control of Eukaryotic Transcription
[q] In the diagram below, the stress is drought. The drought response element is indicated by
[textentry single_char=”true”]
[c]IG M=[Qq]
[f]IE5pY2Ugam9iLiBUaGUgZHJvdWdodCByZXNwb25zZSBlbGVtZW50IGlzIGluZGljYXRlZCBieSAmIzgyMjA7Yy4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJdCYjODIxNztzIHBhcnQgb2YgdGhlIHByb21vdGVyIHJlZ2lvbiBvZiB0aGUgZ2VuZXMuwqA=[Qq]
[q] In the diagram below, an activated transcription factor is represented by
[textentry single_char=”true”]
[c]IG I=[Qq]
[f]IE5pY2Ugam9iISBMZXR0ZXIgJiM4MjIwO2ImIzgyMjE7IGluZGljYXRlcyBhbiBhY3RpdmF0ZWQgdHJhbnNjcmlwdGlvbiBmYWN0b3Iu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgYWN0aXZhdGVkIHRyYW5zY3JpcHRpb24gZmFjdG9yIGNhbiBiaW5kIHdpdGggKG9yIG5lYXIpIHRoZSBwcm9tb3RlciByZWdpb24u[Qq]
[q] In the diagram below, RNA polymerase is represented by
[textentry single_char=”true”]
[c]IG c=[Qq]
[f]IEV4Y2VsbGVudCEgUk5BIHBvbHltZXJhc2UgaXMgcmVwcmVzZW50ZWQgYnkgJiM4MjIwO2cuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgYWN0aXZhdGVkIHRyYW5zY3JpcHRpb24gZmFjdG9yIGNhbiBiaW5kIHdpdGggKG9yIG5lYXIpIHRoZSBwcm9tb3RlciByZWdpb24u[Qq]
[q] In the diagram below, messenger RNA is represented by
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IEV4Y2VsbGVudCEgTWVzc2VuZ2VyIFJOQSBpcyByZXByZXNlbnRlZCBieSAmIzgyMjA7ZS4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBMZXR0ZXIgJiM4MjIwO2cmIzgyMjE7IHJlcHJlc2VudHMgUk5BIHBvbHltZXJhc2UmIzgyMTc7cyByb2xlLiBJdCB0cmFuc2NyaWJlcyBETkEgaW50byBSTkEuIElmICYjODIyMDtnJiM4MjIxOyBpcyBSTkEgcG9seW1lcmFzZSwgdGhlbiBtUk5BIG11c3QgYmUgJiM4MjMwOw==[Qq]
[q] In the diagram below, estrogen would be
[textentry single_char=”true”]
[c]IE Y=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtGJiM4MjIxOyByZXByZXNlbnRzIGVzdHJvZ2VuLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIGhvcm1vbmUgdGhhdCYjODIxNztzIGNvbWluZyBmcm9tIG91dHNpZGUgdGhlIGNlbGwsIHRoZW4gZGlmZnVzaW5nIGludG8gdGhlIGN5dG9wbGFzbSB0aHJvdWdoIHRoZSBjZWxsIG1lbWJyYW5lLg==[Qq]
[q] In the diagram below, a mobile estrogen receptor before it has bound to estrogen would be represented by
[textentry single_char=”true”]
[c]IE c=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtHJiM4MjIxOyByZXByZXNlbnRzIGEgbW9iaWxlIGVzdHJvZ2VuIHJlY2VwdG9yIGJlZm9yZSBpdCBiaW5kcyB3aXRoIGVzdHJvZ2VuLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHJlY2VwdG9yIHRoYXQgY2FuIGJpbmQgd2l0aCBlc3Ryb2dlbiAod2hpY2ggaXMgcmVwcmVzZW50ZWQgYnkgJiM4MjIwO0YuJiM4MjIxOyk=[Qq]
[q] In the diagram below, which letter represents the estrogen-receptor complex acting as a transcription factor?
[textentry single_char=”true”]
[c]IG k=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtJJiM4MjIxOyBzaG93cyBhbiBlc3Ryb2dlbi9yZWNlcHRvciBjb21wbGV4IGludGVyYWN0aW5nIHdpdGggRE5BLCBhY3RpbmcgYXMgYSB0cmFuc2NyaXB0aW9uIGZhY3Rvci4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHJlY2VwdG9yIGJvdW5kIHRvIGVzdHJvZ2VuLCBpbnRlcmFjdGluZyB3aXRoIEROQS4=[Qq]
[q] In the diagram below, which letter represents the part that’s translating the transcription product into protein?
[textentry single_char=”true”]
[c]IE w=[Qq]
[f]IENvcnJlY3QhIExldHRlciAmIzgyMjA7TCYjODIyMTsgc2hvd3MgYSByaWJvc29tZS4gdGhlIHJpYm9zb21lIGlzIHRyYW5zbGF0aW5nIFJOQSAodGhlIHRyYW5zY3JpcHRpb24gcHJvZHVjdCkgaW50byBwcm90ZWlu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgdHJhbnNjcmlwdGlvbiBwcm9kdWN0IGlzIFJOQSAoZmlyc3Qgc2hvd24gYXQgJiM4MjIwO0smIzgyMjE7KS4gV2hhdCYjODIxNztzIHRyYW5zbGF0aW5nIHRoZSBSTkEgaW50byBwcm90ZWluPw==[Qq]
[q] A key idea represented below is that the action of genes can be [hangman] if these genes share common [hangman] sequences.
[c]IGNvb3JkaW5hdGVk[Qq]
[c]IHJlZ3VsYXRvcnk=[Qq]
[x][restart]
[/qwiz]
4. Regulation of translation by microRNAs (RNA interference)
Up to this point, the mechanisms for control of gene expression have occurred at the level of DNA or RNA. Now we’ll look at a regulatory mechanism that occurs during translation. While there are a variety of translation and post-translational control mechanisms, we’re only going to look at one: microRNAs and RNA interference.
MicroRNA’s fall into an important category of RNAs that are called non-coding RNAs. These are RNAs that are not involved in translation (such as mRNA, rRNA, or tRNA) but which play a role in regulating gene expression.
As an AP Biology student, the main type of non-protein-coding RNAs to know about are microRNAs (miRNAs). These miRNAs are about 22 nucleotides long. They originate from longer precursor RNAs that are cut apart by an enzyme. After being cut, each 22 nucleotide fragment forms a complex with a protein.
One of the main functions of miRNA is RNA interference. Using complementary base pairing, the miRNA can bind with messenger RNA. Binding blocks mRNA translation into proteins and is sometimes followed by enzymatic degradation of the mRNA.
In a previous tutorial, we discussed how messenger RNA has to be processed before it can leave the nucleus and be translated into protein. Similarly, microRNAs need to be processed before they can play their role in the cytoplasm (which is regulating translation).
The image on the left illustrates microRNA processing and its subsequent regulatory activities. Letter “A” shows DNA that codes for miRNA. This DNA is transcribed (“B”) into a pre-microRNA (“C”). Processing and export from the nucleus (“D”) convert this into a mature microRNA (“E”), which then associates with a protein called an RNA-induced silencing complex (“I”). If the miRNA partially matches its target messenger RNA (as shown at “H”) then translation is blocked (“J”). A complete match (“G”) results in RNA degradation (“K”). Both cases interrupt the translation of proteins from mRNA, which is why the entire phenomenon is called RNA interference.
What’s the takeaway? RNAi provides cells with a means of regulating gene expression after transcription.
5. Flashcards: Coordinated Control of Gene Expression and MicroRNA
[qdeck style=”min-height: 450px width: 650px !important;” bold_text=”false” qrecord_id=”sciencemusicvideosMeister1961-Coordinated Control of Eukaryotic Gene Expression and MicroRNA FC (v2.0)”]
[h]Coordinated Control of Eukaryotic Gene Expression and MicroRNA
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|180b7bf217510″ question_number=”247″ topic=”6.5.Regulation_of_Gene_Expression”] Using estrogen as an example, explain how gene expression can be coordinated in a variety of tissues.
[a] In eukaryotes, gene expression can be coordinated by having separate genes share regulatory sequences that respond to the same transcription factors. For example, cells that respond to estrogen develop in a way so that in addition to the tissue-specific genes that they express, they also express the gene for the estrogen receptor. As a result, when estrogen (F) is released from the ovaries, it will bind with these cells’ cytoplasmic estrogen receptors (G) and induce transcription (I) of estrogen-related genes.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|18319c34ccd10″ question_number=”241″ topic=”6.5.Regulation_of_Gene_Expression”] Using RNA interference as an example, describe the role of small RNA molecules in regulating gene expression.
[a]One of the main functions of small RNA molecules (typically about 22 nucleotides long) is RNA interference. In RNA interference, small RNAs (E) bind with messenger RNA (I), often blocking the translation of the mRNA into protein (J), and sometimes signaling for that mRNA’s enzymatic destruction (K).
[/qdeck]
6. Checking Understanding: RNA Interference
The quiz below reviews what’s above, as well as some general features of post-transcriptional modification of RNA.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-RNA interference (v2.0)”]
[h] RNA Interference
[q multiple_choice=”true”]
[l]miRNA
[f*] Excellent!
[fx] No. Please try again.
[l]mRNA degradation
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]DNA
[f*] Good!
[fx] No, that’s not correct. Please try again.
[l]precursor miRNA
[f*] Good!
[fx] No. Please try again.
[l]Blocked translation
[f*] Good!
[fx] No. Please try again.
[q multiple_choice=”true”] The type of RNA with a regulatory function is
[c]IG1STkE=[Qq]
[f]IE5vLiBtUk5BIGlzIG1lc3NlbmdlciBSTkEuIEl0IGdldHMgdHJhbnNsYXRlZCBpbnRvIHByb3RlaW4u[Qq]
[c]IG1p Uk5B[Qq]
[f]IENvcnJlY3QuIG1pUk5BIGlzIG1pY3JvUk5BLCBhbmQgaXRzIHJvbGUgaXMgcmVndWxhdGluZyB0cmFuc2xhdGlvbi4=[Qq]
[c]IHRSTkE=[Qq]
[f]IE5vLiB0Uk5BIGlzIHRyYW5zZmVyIFJOQS4gSXRzIHJvbGUgaXMgdG8gYnJpbmcgYW1pbm8gYWNpZHMgdG8gdGhlIHJpYm9zb21lIGR1cmluZyBwcm90ZWluIHN5bnRoZXNpcy4=[Qq]
[q] In the diagram below, the RNA-induced silencing complex is represented by the letter
[textentry single_char=”true”]
[c]IE k=[Qq]
[f]IE5pY2UhICYjODIyMDtJJiM4MjIxOyByZXByZXNlbnRzIHRoZSBSTkEtaW5kdWNlZCBzaWxlbmNpbmcgY29tcGxleC4=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIG1pUk5BIHRoYXQmIzgyMTc7cyBhdHRhY2hlZCB0byBhIHByb3RlaW4sIHdoaWNoIGlzIGluIHR1cm4gYXR0YWNoZWQgdG8gdGhlIHJpYm9zb21lLg==[Qq]
[q] In the diagram below, the RNA-induced silencing complex is represented by the letter
[textentry single_char=”true”]
[c]IE k=[Qq]
[f]IE5pY2UhICYjODIyMDtJJiM4MjIxOyByZXByZXNlbnRzIHRoZSBSTkEtaW5kdWNlZCBzaWxlbmNpbmcgY29tcGxleC4=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIG1pUk5BIHRoYXQmIzgyMTc7cyBhdHRhY2hlZCB0byBhIHByb3RlaW4sIHdoaWNoIGlzIGluIHR1cm4gYXR0YWNoZWQgdG8gdGhlIHJpYm9zb21lLg==[Qq]
[q] In the diagram below, a silenced ribosome is represented by the letter
[textentry single_char=”true”]
[c]IE Y=[Qq]
[f]IEdvb2Qgam9iISAmIzgyMjA7RiYjODIyMTsgcmVwcmVzZW50cyBhIHJpYm9zb21lIHRoYXQmIzgyMTc7cyBiZWVuIHNpbGVuY2VkIGJ5IFJOQSBpbnRlcmZlcmVuY2Uu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJbiB0aGlzIGRpYWdyYW0sIG1STkEgaXMgcmVwcmVzZW50ZWQgYXMgYSBibHVlLCBzaW5nbGUgc3RyYW5kLiBXaGF0JiM4MjE3O3MgJiM4MjIwO3JlYWRpbmcmIzgyMjE7IHRoZSBtUk5BPw==[Qq]
[q]The phenomenon shown below is called [hangman] [hangman].
[c]Uk5B[Qq]
[c]aW50ZXJmZXJlbmNl[Qq]
[q]The molecule shown at “E” is called an [hangman] (use an abbreviation, like “tRNA”).
[c]bWlSTkE=[Qq]
[q] In the diagram below, mature miRNA is represented by the letter
[textentry single_char=”true”]
[c]IE U=[Qq]
[f]IEF3ZXNvbWUhICYjODIyMDtFJiM4MjIxOyByZXByZXNlbnRzIGEgbWF0dXJlIG1pUk5BLCByZWFkeSB0byBmb3JtIGFuIFJOQS1pbmR1Y2VkIHNpbGVuY2luZyBjb21wbGV4IGFuZCBibG9jayB0cmFuc2xhdGlvbi4=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJbiB0aGlzIGRpYWdyYW0sIHByZS1taVJOQSBpcyByZXByZXNlbnRlZCBieSB0aGUgbGV0dGVyICYjODIyMDtDLiYjODIyMTsgV2hpY2ggbGV0dGVyIHdvdWxkIHNob3cgYSBwcm9jZXNzZWQgZm9ybSBvZiAmIzgyMjA7Qz8mIzgyMjE7[Qq]
[/qwiz]
7. What’s next?
Proceed to Topics 6.5 – 6.6, Part 5: Eukaryotic Gene Regulation and Development (the next tutorial in AP Bio Unit 6)