Click here for the Molecules of Life Student Learning Guide
Page Outline
- The Functions of Proteins
- Proteins are Polymers of Amino Acids, Linked by Peptide Bonds
- Amino Acids, Peptide Bonds, and Polypeptide Quiz
- Protein shape
- Protein Shape and Genes
- Understanding Protein Folding
- Protein shape quiz
- Denaturation
- Proteins quiz
1. The Functions of Proteins
Let’s start our study of proteins by considering our hands.

Our fingernails are composed of a protein called keratin. A different form of keratin makes up the outermost layer of your skin. The elasticity of deeper layers of your skin — your skin’s ability to return to its shape after being pinched or stretched — is made possible by a protein called elastin.
Go a bit deeper. Your fingers move because they’re attached to muscles by tendons and ligaments that are composed of a protein called collagen. The muscles that move the bones in your hand are made of proteins such as actin and myosin.
Think about your veins. The red blood cells that flow within them are filled with an oxygen carrying protein called hemoglobin. If you were to cut your hand and get an infection, your immune system would produce proteins called immunoglobulins to form the antibodies that would beat back the invading bacteria.
When you look at any animal, you’re looking at protein. The same is not true of plants, which are mostly carbohydrate. But how did that carbohydrate get there? The enzyme that pulls carbon dioxide out of the air to create carbohydrates during photosynthesis is called RuBisCo. Like almost all enzymes, it’s a protein.
I could go on. But let’s organize our thinking about the various functions of proteins by completing the interactive table below. Use prior knowledge and trial and error (and don’t worry about getting every answer right the first time).
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Functions of proteins (HS)”]
[h]Some of the functions of proteins
[q labels = “top”]
Proteins
- Control chemical reactions as ______________.
- Create _____________ like bone, hair, feathers (keratin)
- Fight __________ (antibodies)
- Produce movement (___________)
- ____________ oxygen (hemoglobin in red blood cells)
- Store __________________ (albumin in egg white)
- Transmit _________ as hormones and neurotransmitters.
PROTEINS ARE THE ___________________ MACROMOLECULES
[l]chemical energy
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]infection
[fx] No. Please try again.
[f*] Correct!
[l]enzymes
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]MOST DIVERSE
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]muscle
[fx] No. Please try again.
[f*] Correct!
[l]structures
[fx] No. Please try again.
[f*] Excellent!
[l]transport
[fx] No. Please try again.
[f*] Correct!
[l]signals
[fx] No. Please try again.
[f*] Good!
[/qwiz]
2. Proteins are polymers of amino acids, linked by peptide bonds
2a. Amino Acids
A key idea of this unit is that many of the molecules of life are polymers composed of monomers. For example, polysaccharides like starch and cellulose are polymers of monosaccharides. For proteins (which are themselves polymers), the monomers are amino acids.
Twenty amino acids build all the proteins found in living things. Each amino acid has three letter abbreviation: “lys” for lysine, “ser” for serine, etc.
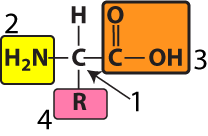
All amino acids are variations of a common structure. At the center of every amino acid is a central carbon atom (shown at “1” in the diagram to your left). The central carbon is also referred to as the alpha (α) carbon. This central carbon is bonded to four things: a hydrogen atom, an —NH2 on one side, and a —COOH on the other side. The —NH2 is called an “amino group,” and the —COOH is called a “carboxylic acid.” That’s where the name “amino acid” comes from.
The fourth bond connects to what’s called an “R group” or a “side chain.” This is shown at number “4.” Each of the twenty amino acids has a distinct R group. The R groups are shaded in the diagram below. Just take a look at the twenty amino acids below, noting how their R groups are different.
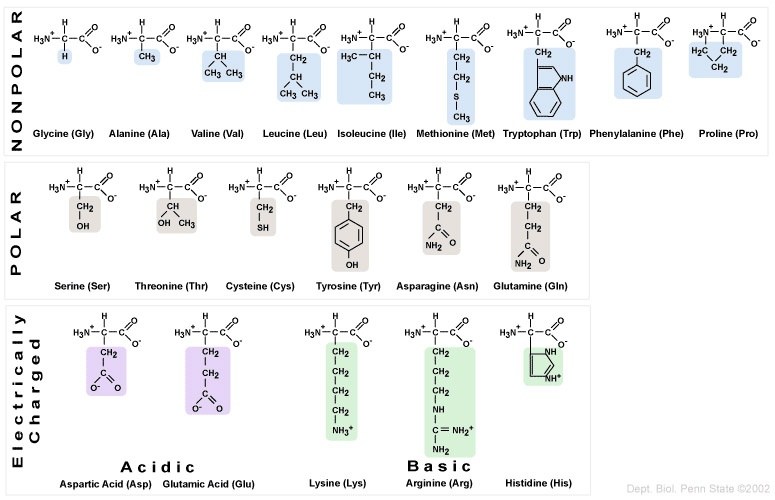
The twenty amino acids can be organized into four groups, based on the chemistry of their side chains. These R groups can be
- Nonpolar/hydrophobic
- Polar/hydrophilic
- Acidic, or negatively charged.
- Basic, or positively charged.
The main thing to know is that amino acids have different chemical properties, based on their side chains. This is important because mutations in DNA that offspring inherit from their parents can result in substitutions of one amino acid for another. If the substituted amino acid has a different chemistry from the original amino acid, then the effect on the protein’s structure and function can be significant.
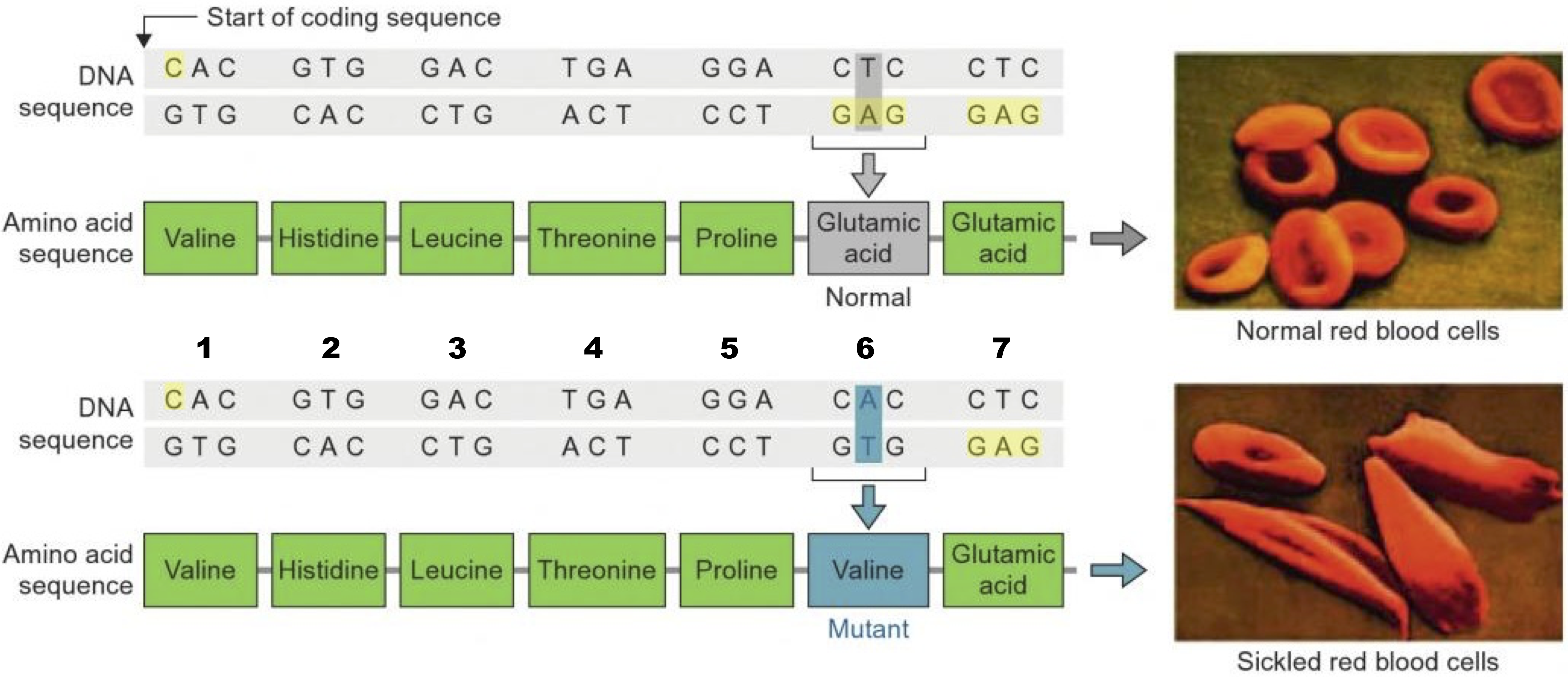 For example, the inherited blood disorder sickle cell anemia is caused by a substitution of a nonpolar acidic amino acid for an acidic amino acid.
For example, the inherited blood disorder sickle cell anemia is caused by a substitution of a nonpolar acidic amino acid for an acidic amino acid.
Sickle cell anemia is the most common inherited genetic disease among Americans of African descent. People with the disease experience crippling pain crises. These crises are caused when blood oxygen levels get low, which can happen whenever a person exerts themself (by exercising, for example).
What causes these pain crises? It has to do with a protein called hemoglobin, which carries oxygen in red blood cells. Hemoglobin is made of dozens of amino acids, Seven of these amino acids are shown in the chart above.
In people with sickle cell anemia, the DNA they inherited from their parents codes for the non-polar amino acid valine instead of glutamic acid (which is acidic). The substitution changes hemoglobin’s chemistry so that hemoglobin molecules stick together in low oxygen conditions. That causes them to form rods, which cause red blood cells to become spiked instead of smooth (see the photos above). The spiked cells clump together, blocking capillaries. These blocked capillaries keep blood from flowing to the tissues, causing extreme pain (and tissue damage).
What’s the takeaway: a major genetic disease, affecting millions of people, is caused by substituting one type of amino acid for another.
At the same time, it’s important to note that mutations that cause changes in amino acids aren’t always bad. In fact, one major mechanism of evolution is mutations in DNA that change the structure of proteins in beneficial ways. We’ll learn more about that when we learn about evolution later in this course.
2.b. Peptide Bonds
How are proteins made? They’re assembled by cells During the a process that’s called protein synthesis.
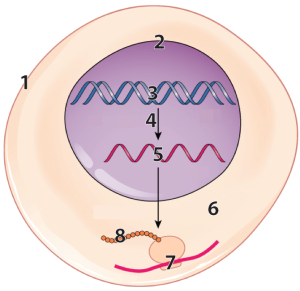
The diagram at left represents a cell, with some parts vastly enlarged to represent the information flow that enables cells to make protein. Number “1” represents the cell’s outer boundary, or membrane.
DNA (number “3”) in the cell’s nucleus (“2”) has instructions for making a molecule called RNA (“5”).
That RNA is synthesized through a process called transcription, which is indicated by the arrow at “4.” After transcription, a variety of processes in the nucleus modify the RNA, converting it into messenger RNA (mRNA).
The mRNA leaves the nucleus and enters the cytoplasm (“6”), where tiny particles called ribosomes (“7”) translate the RNA into a protein (“8”).
Lets zoom in on a ribosome (“2, below) to see a bit more about how protein synthesis works. In the diagram below, messenger RNA is shown at “3.” Following the instructions in the mRNA, ribosomes link amino acids (“1”) together. .
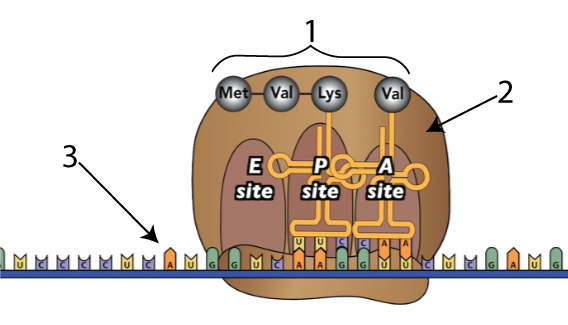
Chemically, here’s what happens when ribosomes link amino acids together.
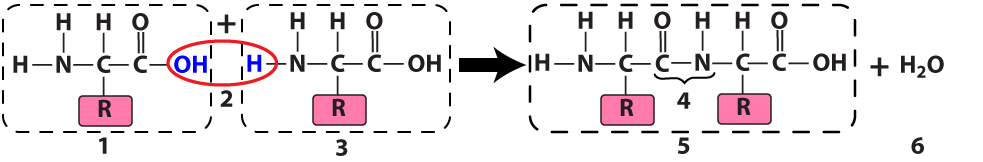
The ribosome connects two amino acids (“1” and “3”) by removing an —OH from one amino acid and an —H (a hydrogen atom) from another. In the diagram above, the result is a dipeptide (“5”): two amino acids linked by a peptide bond (at “4”). Because this is a dehydration synthesis, a water molecule (at “6”) also results from this reaction.
2c. Polypeptides
As the ribosome adds more amino acids, the result is a polypeptide (a chain of linked amino acids).
In the same way as the letters in a word can be in any order, the amino acids in a protein can be in any order. As I’ve noted previously, however, the “letters to words = amino acids to protein” analogy is limited. Proteins can be thousands of amino acids long, making the universe of potential proteins much greater than the universe of potential words.
3. Amino Acids, Peptide Bonds, and Polypeptides: Checking Understanding
Just to keep your previous learning intact, the quiz below includes some review questions about the molecules of life.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-amino acids and polypeptides (HS)”]
[h] Proteins Quiz 1: Amino Acids, Peptide Bonds, Polypeptides
[q] The monomers of carbohydrates are
[c]IGFtaW5vIGFjaWRzLg==[Qq]
[f]IE5vLiBBbWlubyBhY2lkcyBhcmUgdGhlIG1vbm9tZXJzIG9mIHByb3RlaW5zLg==[Qq]
[c]IGZhdHR5IGFjaWRzLg==[Qq]
[f]IE5vLiBGYXR0eSBhY2lkcyBhcmUgYSBrZXkgYnVpbGRpbmcgYmxvY2sgb2YgbWFueSBsaXBpZHMu[Qq]
[c]IG1vbm9zYWNj aGFyaWRlcy4=[Qq]
[f]IENvcnJlY3QhIE1vbm9zYWNjaGFyaWRlcyBhcmUgdGhlIG1vbm9tZXJzIG9mIGNhcmJvaHlkcmF0ZXM=[Qq]
[c]IG51Y2xlb3RpZGVzLg==[Qq]
[f]IE5vLiBOdWNsZW90aWRlcyBhcmUgdGhlIG1vbm9tZXJzIG9mIG51Y2xlaWMgYWNpZHMgKGxpa2UgRE5BIGFuZCBSTkEp[Qq]
[q json=”true” multiple_choice=”true” xx=”2″ dataset_id=”SMV_biochem_7_amino acids and peptide bonds|e9225fb4e63fa” question_number=”3″] The monomers of proteins are
[c]IGFtaW5vIG FjaWRzLg==[Qq]
[f]IEV4Y2VsbGVudC4gQW1pbm8gYWNpZHMgYXJlIHRoZSBtb25vbWVycyBvZiBwcm90ZWlucy4=[Qq]
[c]IGZhdHR5IGFjaWRzLg==[Qq]
[f]IE5vLiBGYXR0eSBhY2lkcyBhcmUgYSBrZXkgYnVpbGRpbmcgYmxvY2sgb2YgbWFueSBsaXBpZHMu[Qq]
[c]IG1vbm9zYWNjaGFyaWRlcy4=[Qq]
[f]IE5vLiBNb25vc2FjY2hhcmlkZXMgYXJlIHRoZSBtb25vbWVycyBvZiBjYXJib2h5ZHJhdGVz[Qq]
[c]IG51Y2xlb3RpZGVzLg==[Qq]
[f]IE5vLiBOdWNsZW90aWRlcyBhcmUgdGhlIG1vbm9tZXJzIG9mIG51Y2xlaWMgYWNpZHMgKGxpa2UgRE5BIGFuZCBSTkEp[Qq]
[q json=”true” multiple_choice=”true” xx=”2″ dataset_id=”SMV_biochem_7_amino acids and peptide bonds|e91e4ca0173fa” question_number=”4″] In the diagram below, which number indicates the central carbon atom?
[c]ID E=[Qq]
[f]IEV4Y2VsbGVudCEgJiM4MjIwOzEmIzgyMjE7IGlzIHRoZSBjZW50cmFsIGNhcmJvbg==[Qq]
[c]IDI=[Qq]
[f]IE5vLiAmIzgyMjA7MiYjODIyMTsgaXMgYW4gYW1pbm8gZ3JvdXAu[Qq]
[c]IDM=[Qq]
[f]IE5vLiAmIzgyMjA7MyYjODIyMTsgaXMgdGhlIGNhcmJveHlsaWMgYWNpZA==[Qq]
[c]IDQ=[Qq]
[f]IE5vLiAmIzgyMjA7NCYjODIyMTsgaXMgdGhlICYjODIyMDtSJiM4MjIxOyBncm91cC4gSXQmIzgyMTc7cyB0aGUgcGFydCB0aGF0IHZhcmllcyBmcm9tIG9uZSBhbWlubyBhY2lkIHRvIHRoZSBuZXh0Lg==[Qq]
[q json=”true” multiple_choice=”true” xx=”2″ dataset_id=”SMV_biochem_7_amino acids and peptide bonds|e91a144a89ffa” question_number=”5″] The “amino” part of “amino acids” refers to which part of the diagram below?
[c]IDE=[Qq]
[f]IE5vICYjODIyMDsxJiM4MjIxOyBpcyB0aGUgY2VudHJhbCBjYXJib24=[Qq]
[c]ID I=[Qq]
[f]IEdyZWF0IGpvYi4gJiM4MjIwOzImIzgyMjE7IGlzIGFuIGFtaW5vIGdyb3VwLg==[Qq]
[c]IDM=[Qq]
[f]IE5vLiAmIzgyMjA7MyYjODIyMTsgaXMgdGhlIGNhcmJveHlsaWMgYWNpZA==[Qq]
[c]IDQ=[Qq]
[f]IE5vLiAmIzgyMjA7NCYjODIyMTsgaXMgdGhlICYjODIyMDtSIGdyb3VwJiM4MjIxOyBvciAmIzgyMjA7c2lkZSBjaGFpbi4mIzgyMjE7[Qq]
[q json=”true” multiple_choice=”true” xx=”2″ dataset_id=”SMV_biochem_7_amino acids and peptide bonds|e9160135baffa” question_number=”6″] In the diagram below, which number indicates the part that makes an amino acid an acid?
[c]IDE=[Qq]
[f]IE5vLiAmIzgyMjA7MSYjODIyMTsgaXMgdGhlIGNlbnRyYWwgY2FyYm9u[Qq]
[c]IDI=[Qq]
[f]IE5vLiAmIzgyMjA7MiYjODIyMTsgaXMgYW4gYW1pbm8gZ3JvdXAu[Qq]
[c]ID M=[Qq]
[f]IFdheSB0byBnbyEgJiM4MjIwOzMmIzgyMjE7IGlzIHRoZSBjYXJib3h5bGljIGFjaWQ=[Qq]
[c]IDQ=[Qq]
[f]IE5vLiAmIzgyMjA7NCYjODIyMTsgaXMgdGhlICYjODIyMDtSIGdyb3VwJiM4MjIxOyBvciAmIzgyMjA7c2lkZSBjaGFpbi4mIzgyMjE7[Qq]
[q json=”true” multiple_choice=”true” xx=”2″ dataset_id=”SMV_biochem_7_amino acids and peptide bonds|e911c8e02dbfa” question_number=”7″] In the diagram below, which number indicates the part that varies from one amino acid to the next?
[c]IDE=[Qq]
[f]IE5vLiAmIzgyMjA7MSYjODIyMTsgaXMgdGhlIGNlbnRyYWwgY2FyYm9uLCB3aGljaCBpcyB0aGUgc2FtZSBpbiBldmVyeSBhbWlubyBhY2lkLg==[Qq]
[c]IDI=[Qq]
[f]IE5vLiAmIzgyMjA7MiYjODIyMTsgaXMgYW4gYW1pbm8gZ3JvdXAsIHdoaWNoIGlzIHRoZSBzYW1lIGluIGV2ZXJ5IGFtaW5vIGFjaWQu[Qq]
[c]IDM=[Qq]
[f]IE5vICYjODIyMDszJiM4MjIxOyBpcyB0aGUgY2FyYm94eWxpYyBhY2lkLCB3aGljaCBpcyB0aGUgc2FtZSBpbiBldmVyeSBhbWlubyBhY2lkLg==[Qq]
[c]ID Q=[Qq]
[f]IFllcyEgJiM4MjIwOzQmIzgyMjE7IGlzIHRoZSAmIzgyMjA7UiBncm91cCwmIzgyMjE7IHRoZSBwYXJ0IHRoYXQgY2FuIHZhcnkgZnJvbSBvbmUgYW1pbm8gYWNpZCB0byB0aGUgbmV4dC4=[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_7_amino acids and peptide bonds|e90d908aa07fa” question_number=”8″] In the diagram below, which number indicates a peptide bond?
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IEV4Y2VsbGVudCEgJiM4MjIwOzQmIzgyMjE7IGlzIHBvaW50aW5nIHRvIHRoZSBwZXB0aWRlIGJvbmQu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIHRoZSBib25kIHRoYXQgY29ubmVjdHMgdGhlIGFtaW5vIGFjaWRzIGF0ICYjODIyMDsxJiM4MjIxOyBhbmQgJiM4MjIwOzMmIzgyMjE7ICh0aGUgcmVhY3RhbnRzKSBpbiB0aGUgZGlwZXB0aWRlIGF0ICYjODIyMDs1JiM4MjIxOyAodGhlIHByb2R1Y3QpLiBSZW1lbWJlciB0aGF0IHBlcHRpZGUgYm9uZHMgaW52b2x2ZSB0aGUgcmVtb3ZhbCBvZiBhbiAmIzgyMjA7SCYjODIyMTsgYW5kIGFuICYjODIyMDstT0gmIzgyMjE7IHRvIGZvcm0gYSB3YXRlciBtb2xlY3VsZS4=[Qq]
[q] In the diagram below, a newly made protein is shown at
[textentry single_char=”true”]
[c]ID g=
[f]IE5pY2UhIE51bWJlciAmIzgyMjA7OCYjODIyMTsgaW5kaWNhdGVzIGEgbmV3bHkgbWFkZSBwcm90ZWluLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIGEgcmlib3NvbWUuIEl0JiM4MjE3O3Mgc29tZXRoaW5nIHRoYXQgdGhlIHJpYm9zb21lIGlzIG1ha2luZy4=[Qq]
[q] In the diagram below, the inherited instructions for making protein are indicated by number _____, which represents DNA.
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IEdvb2Qgd29yayEgTnVtYmVyICYjODIyMDszJiM4MjIxOyBpbmRpY2F0ZXMgRE5BLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBZb3UmIzgyMTc7cmUgbG9va2luZyBmb3IgRE5BLCB3aGljaCBpcyB0aGUgZ2VuZXRpYyBtYXRlcmlhbCB5b3UgaW5oZXJpdCBmcm9tIHlvdXIgcGFyZW50cy4=[Qq]
[q] In the diagram below, what number represents a ribosome?
[textentry single_char=”true”][c]ID I=
Cg==[Qq][f]IEV4Y2VsbGVudC4gQSByaWJvc29tZSBpcyBzaG93biBhdCAmIzgyMjA7Mi4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBBIHJpYm9zb21lIGlzIGEgcGFydGljbGUgdGhhdCBtYWtlcyBwcm90ZWluLiBJZiAmIzgyMjA7MSYjODIyMTsgc2hvd3MgYSBncm93aW5nIHByb3RlaW4sIHdoYXQgcGFydCBjb3VsZCBiZSBtYWtpbmcgdGhhdCBwcm90ZWluPw==[Qq]
[q] In the diagram below, a growing protein is represented by number
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IEdvb2Qgd29yay4gQSBncm93aW5nIHByb3RlaW4gaXMgcmVwcmVzZW50ZWQgYnkgJiM4MjIwOzEuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGlzIGdyb3dpbmcgcHJvdGVpbiBjb25zaXN0cyBvZiAzIGFtaW5vIGFpZHMsIGFuZCBhIDR0aCBpcyBhYm91dCB0byBiZSBhZGRlZCB0byBpdC4=[Qq]
[q]The diagram below represents how during [hangman] synthesis, a [hangman] at “2” reads instructions that come in the form of messenger [hangman], shown at “3.”
[c]cHJvdGVpbg==[Qq]
[c]cmlib3NvbWU=[Qq]
[c]Uk5B[Qq]
[q]The diagram below shows how a single [hangman] in DNA can result in a substitution of the [hangman] [hangman] valine for glutamic acid. The result is the inherited genetic disease, sickle cell [hangman].
[c]bXV0YXRpb24=[Qq]
[c]YW1pbm8=[Qq]
[c]YWNpZA==[Qq]
[c]YW5lbWlh[Qq]
[/qwiz]
4. Proteins Have Specific Shapes
One of the most remarkable features of proteins is how specific their shape is. To get an understanding of this, let’s think about our immune system.
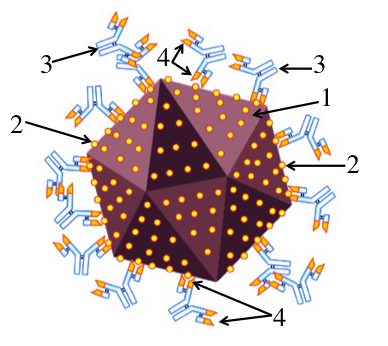
Imagine that you’ve been infected by a virus (“1”). Your immune system counterattacks by producing proteins called antibodies (“3”) that can bind with specific molecules on the surface of the virus. These molecules are called antigens (“2”).
An antibody can bind with an antigen because the tip of the antibody has an antigen binding site (“4”) whose shape closely matches with, or complements, the shape of the antigen. This binding either directly neutralizes the virus (keeping it, for example, from infecting a cell), or allows other parts of the immune system to neutralize and destroy the virus.
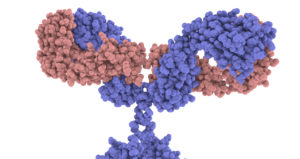
In the diagram above, I drew the antigen as a circle, and gave the antigen binding site on the antibody a complementary shape. But to get a sense of the required specificity of the antigen binding site, take a look at the image at right (from the Garvan Institute of Medical Research / Centre for Targeted Therapy). This is a space filling model, with every atom represented. Please click it to take a close look at the antigen binding sites on the tips of the arms.
The key point is that this shape, down to the nanometer scale, is specific. It can only do its job (binding with and neutralizing a virus) if it has this exact shape. And this leads to the question: how can protein shape be determined so specifically?
5. Protein shape is determined by genes that specify a sequence of amino acids
The specific shape of a protein emerges from three or four levels of molecular interactions, usually referred to as levels of structure.
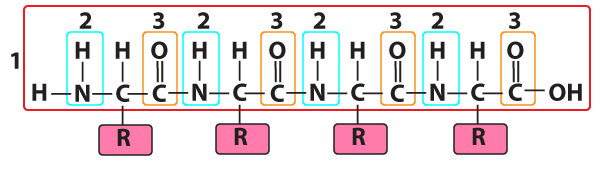
We’ve already discussed the first level. That level is called primary structure. Primary structure is the genetically determined linear sequence of amino acids that makes up a protein.

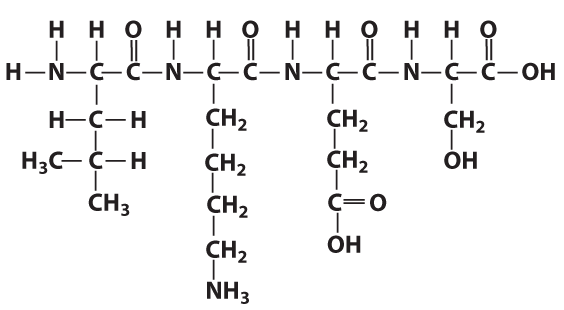
The structural formula on the right shows four linked amino acids. That’s primary structure.
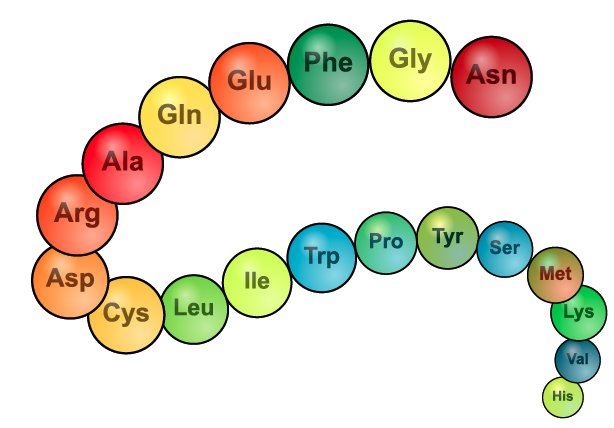 The image on the left is another representation of primary structure. Instead of showing the structural formula, it lists the 3-letter abbreviation for 19 amino acids.
The image on the left is another representation of primary structure. Instead of showing the structural formula, it lists the 3-letter abbreviation for 19 amino acids.
6. Interactions between amino acids cause a protein to fold up into a specific shape
 So, now we need to explain how we get from primary structure — the genetically determined linear sequence of amino acids in a polypeptide chain — to a protein’s three dimensional shape. As you saw above with antibodies, that shape can be very complex. Take a moment to look at this representation of hemoglobin, the oxygen-carrying molecule that we discussed up at the of this page. It’s the opposite of a blob, or a tangle. Every spiral and every hairpin turn is a genetically determined part of hemoglobin’s structure. Change that structure — which as we saw above is what happens in sickle cell anemia — and the result can be disastrous.
So, now we need to explain how we get from primary structure — the genetically determined linear sequence of amino acids in a polypeptide chain — to a protein’s three dimensional shape. As you saw above with antibodies, that shape can be very complex. Take a moment to look at this representation of hemoglobin, the oxygen-carrying molecule that we discussed up at the of this page. It’s the opposite of a blob, or a tangle. Every spiral and every hairpin turn is a genetically determined part of hemoglobin’s structure. Change that structure — which as we saw above is what happens in sickle cell anemia — and the result can be disastrous.
Taking a protein’s primary structure and using it to understand the precise three-dimensional shape of a protein involves some very complex physics and chemistry. In fact, predicting a protein’s shape from its primary structure has been a longstanding problem in biology. Recent progress has been made using supercomputers like DeepMind (you can read about that in Nature magazine). There’s even been a crowd-sourced effort to solve this problem using a program called Foldit.
For most high school level biology courses, knowing that the primary structure of a protein determines its 3-D shape is about all that you need to understand. However, if your course requires more more detail, then read below. If you want an even deeper understanding you can dig into this AP/College level tutorial about protein structure.
6.1. Secondary Structure involves helices and pleated sheets
A protein’s secondary structure involves hydrogen bonds that form between —C=O groups groups and —NH groups within what’s called the “backbone” of a polypeptide. To see this, take a look at the diagram below:
Number “1” is the polypeptide backbone. In the same way that your ribs are attached to your backbone, the side chains/R groups of each amino acid residue are attached to the polypeptide backbone.
The —NH groups have a partially positive charge. The —C=O has a partially negative charge. That means the these two groups, if they get close enough to one another, can form a hydrogen bond. And while hydrogen bonds are relatively weak, they can twist the polypeptide backbone into particular shapes. The two shapes are called an alpha helix, and a beta pleated sheet.
6.1.a. Alpha Helix
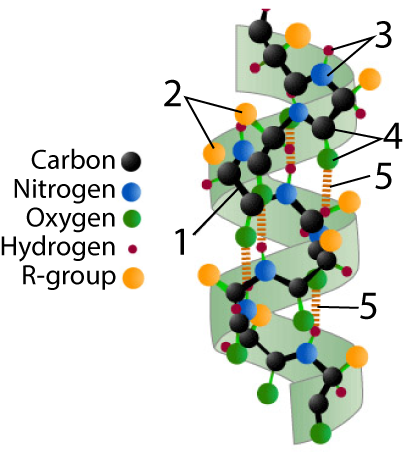
An alpha helix is a corkscrew, stabilized by hydrogen bonds.
At right, you see one such helix. The polypeptide backbone is indicated by “1.” You should be able to notice the repeating pattern of carbon – carbon – nitrogen within the backbone.
The hydrogen bonds that stabilize this shape are shown at “5.”
6.1.b. Beta Pleated Sheet
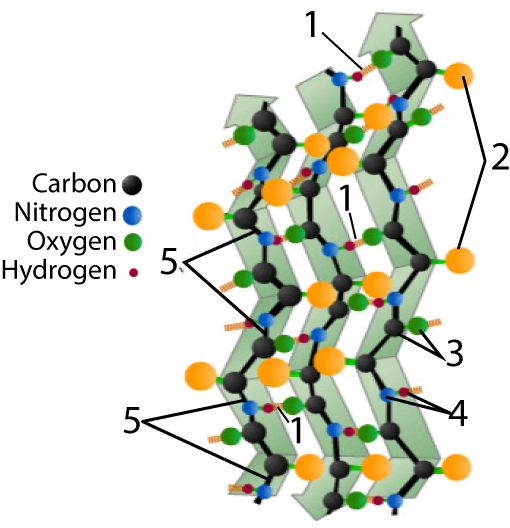 The other secondary structure to know is a beta pleated sheet.
The other secondary structure to know is a beta pleated sheet.
6.2. Tertiary Structure involves interactions between R-groups.
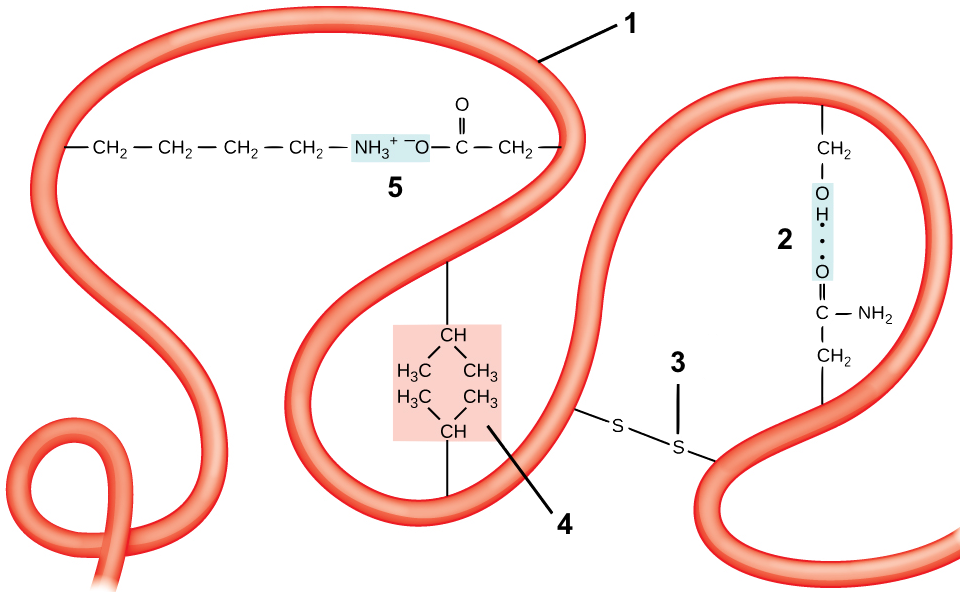
Tertiary structure comes about through interactions between R groups, which cause a polypeptide chain (“1”) to twist and bend. These interactions include
- Hydrogen bonds, shown at “2.”
- Disulfide Bridges. The amino acid cysteine has an —SH at the end of its side chain. If two of these —SH groups in the same polypeptide chain get close to one another, they can form a what’s called a disulfide bridge. A disulfide bridge is a covalent bond.
- Hydrophobic interactions. If side chains that are hydrophobic get close to one another, they can form what’s called a hydrophobic cluster. One such hydrophobic cluster is shown at “4.”
- Ionic bonds. These form when side chains with full positive charges come into proximity with side chains that have full negative charges, as is shown at “5.”
6.4. Quaternary Structure
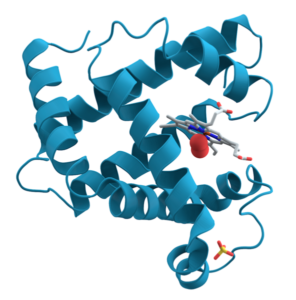
For some proteins, primary, secondary, and tertiary interactions explain the protein’s form and function. Myoglobin, for example, is a protein that stores and releases oxygen within muscle tissue. It consists of 154 amino acids that twist, turn, and coil into the shape shown at right. As you look, I’m hoping that you’re noticing the alpha helices (and connecting them to secondary structure) and hairpin turns (and thinking that some tertiary bond must be causing the polypeptide chain to turn at such a sharp angle).
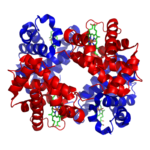
In other proteins, there’s a fourth level of structure, called quaternary. Quaternary structure involves two or more folded polypeptide chains interacting to form a more complex structure.
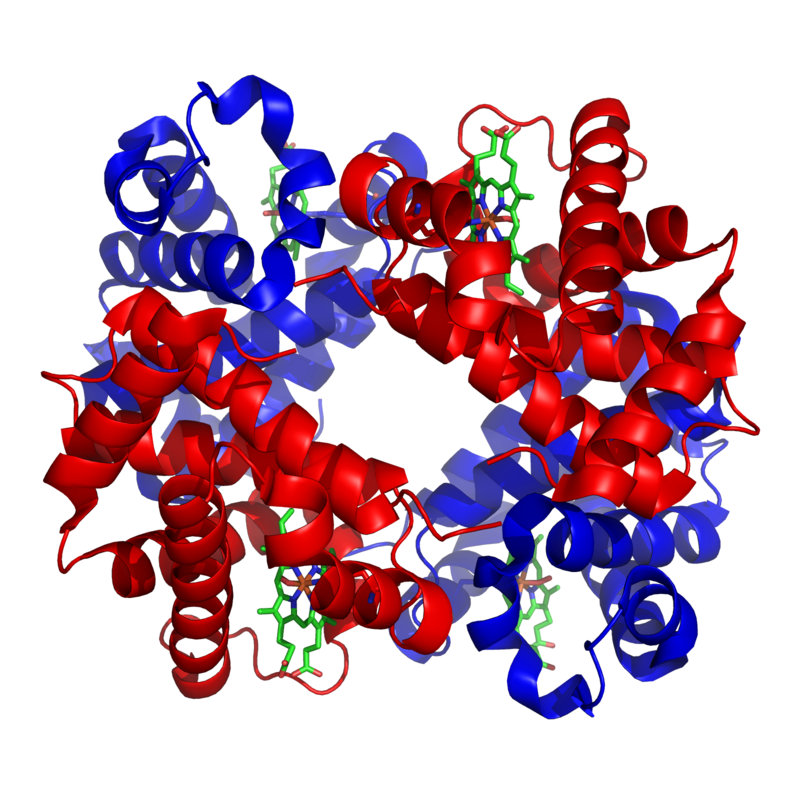
An example of one such quaternary-level protein is hemoglobin, which we’ve discussed several times above. Hemoglobin is the molecule that carries oxygen in our red blood cells. It consists of four polypeptide chains, two are shown in red) and two are shown in blue).
The bonds between the polypeptide subunits in a quaternary protein can include any of the bonds that appear at the tertiary level: hydrogen bonds, hydrophobic interactions, disulfide bridges, and ionic bonds.
7. Protein Shape: Checking Understanding
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Protein shape (HS)”]
[h]Protein Shape: Checking Understanding
[i]
[q] The key point about the diagram below is that antibodies can only neutralize viruses because they have a very [hangman] shape that matches the shape of molecules on a virus’ surface.
[c]IHNwZWNpZmlj[Qq]
[f]IEdvb2Qh[Qq]
[q] The linear sequence of amino acids in a protein is also known as its [hangman] structure .
[c]IHByaW1hcnk=[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] In the diagram below, the spheres with three letter abbreviations represent [hangman] acids, which are the [hangman] of proteins.
[c]IGFtaW5v[Qq]
[f]IEdvb2Qh[Qq]
[c]IG1vbm9tZXJz[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] The bond shown at “5” below is a [hangman] bond. The overall shape that these bonds create within proteins is referred to as an alpha [hangman].
[c]IGh5ZHJvZ2Vu[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IGhlbGl4[Qq]
[f]IENvcnJlY3Qh[Qq]
[q multiple_choice=”true”] In the diagram below, primary structure is indicated by
[c]IG M=[Qq]
[f]IEV4Y2VsbGVudC4gTGV0dGVyICYjODIyMDtjJiM4MjIxOyByZXByZXNlbnRzIHByaW1hcnkgc3RydWN0dXJlLg==[Qq]
[c]IGQ=[Qq]
[f]IE5vLiBUaGUgcHJpbWFyeSBzdHJ1Y3R1cmUgaXMgdGhlIGxpbmVhciBzZXF1ZW5jZSBvZiBhbWlubyBhY2lkcy4=[Qq]
[c]IGo=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO0omIzgyMjE7IGlzIHNob3dpbmcgdGhlIHRoaXJkIGxldmVsICh0ZXJ0aWFyeSBsZXZlbCkgb2YgcHJvdGVpbiBzdHJ1Y3R1cmUuIFRoZSBwcmltYXJ5IHN0cnVjdHVyZSBpcyB0aGUgbGluZWFyIHNlcXVlbmNlIG9mIGFtaW5vIGFjaWRzLg==[Qq]
[c]IGs=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO0smIzgyMjE7IGlzIHNob3dpbmcgNHRoIGxldmVsLCBvciBxdWF0ZXJuYXJ5IHN0cnVjdHVyZS4gUHJpbWFyeSBzdHJ1Y3R1cmUgaXMgdGhlIGxpbmVhciBzZXF1ZW5jZSBvZiBhbWlubyBhY2lkcy4=[Qq]
[q multiple_choice=”true”] In the diagram below, secondary structure is indicated by
[c]IGM=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO2MmIzgyMjE7IHJlcHJlc2VudHMgcHJpbWFyeSBzdHJ1Y3R1cmUuIFNlY29uZGFyeSBzdHJ1Y3R1cmUgaW52b2x2ZXMgaGVsaWNlcyBhbmQgcGxlYXRlZCBzaGVldHMu[Qq]
[c]IG Q=[Qq]
[f]IEZhYnVsb3VzLiBMZXR0ZXIgJiM4MjIwO2QmIzgyMjE7IHJlcHJlc2VudHMgc2Vjb25kYXJ5IHN0cnVjdHVyZS4=[Qq]
[c]IGo=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO0omIzgyMjE7IGlzIHNob3dpbmcgdGhlIHRoaXJkIGxldmVsICh0ZXJ0aWFyeSBsZXZlbCkgb2YgcHJvdGVpbiBzdHJ1Y3R1cmUuIFNlY29uZGFyeSBzdHJ1Y3R1cmUgaW52b2x2ZXMgaGVsaWNlcyBhbmQgcGxlYXRlZCBzaGVldHMu[Qq]
[c]IGs=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO0smIzgyMjE7IGlzIHNob3dpbmcgNHRoIGxldmVsLCBvciBxdWF0ZXJuYXJ5IHN0cnVjdHVyZS4gU2Vjb25kYXJ5IHN0cnVjdHVyZSBpbnZvbHZlcyBoZWxpY2VzIGFuZCBwbGVhdGVkIHNoZWV0cy4=[Qq]
[q multiple_choice=”true”] In the diagram below, tertiary or 3rd level structure is indicated by
[c]IGM=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO2MmIzgyMjE7IHJlcHJlc2VudHMgcHJpbWFyeSBzdHJ1Y3R1cmUuIDNyZCBsZXZlbCBvciB0ZXJ0aWFyeSBzdHJ1Y3R1cmUgaW52b2x2ZXMgdGhlIGNvbXBsZXggYmVuZHMgYW5kIGZvbGRzIHRoYXQgY3JlYXRlIGEgcHJvdGVpbiYjODIxNztzIGZpbmFsIHNoYXBlLg==[Qq]
[c]IGQ=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO2QmIzgyMjE7IHJlcHJlc2VudHMgc2Vjb25kYXJ5IHN0cnVjdHVyZS7CoCAzcmQgbGV2ZWwgb3IgdGVydGlhcnkgc3RydWN0dXJlIGludm9sdmVzIHRoZSBjb21wbGV4IGJlbmRzIGFuZCBmb2xkcyB0aGF0IGNyZWF0ZSBhIHByb3RlaW4mIzgyMTc7cyBmaW5hbCBzaGFwZS4=[Qq]
[c]IG o=[Qq]
[f]IE5pY2UhIExldHRlciAmIzgyMjA7SiYjODIyMTsgaXMgc2hvd2luZyB0aGUgdGhpcmQgbGV2ZWwgKHRlcnRpYXJ5IGxldmVsKSBvZiBwcm90ZWluIHN0cnVjdHVyZS4=[Qq]
[c]IGs=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO0smIzgyMjE7IGlzIHNob3dpbmcgNHRoIGxldmVsLCBvciBxdWF0ZXJuYXJ5IHN0cnVjdHVyZS4gM3JkIGxldmVsIG9yIHRlcnRpYXJ5IHN0cnVjdHVyZSBpbnZvbHZlcyB0aGUgY29tcGxleCBiZW5kcyBhbmQgZm9sZHMgdGhhdCBjcmVhdGUgYSBwcm90ZWluJiM4MjE3O3MgZmluYWwgc2hhcGUu[Qq]
[q multiple_choice=”true”] In the diagram below, 4th level or quaternary structure is indicated by
[c]IGM=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO2MmIzgyMjE7IHJlcHJlc2VudHMgcHJpbWFyeSBzdHJ1Y3R1cmUuIDR0aCBsZXZlbCBvciBxdWF0ZXJuYXJ5IHN0cnVjdHVyZSBpbnZvbHZlcyBib25kcyBiZXR3ZWVuIG11bHRpcGxlLCBmb2xkZWQgcG9seXBlcHRpZGUgY2hhaW5zLg==[Qq]
[c]IGQ=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO2QmIzgyMjE7IHJlcHJlc2VudHMgc2Vjb25kYXJ5IHN0cnVjdHVyZS7CoCA0dGggbGV2ZWwgb3IgcXVhdGVybmFyeSBzdHJ1Y3R1cmUgaW52b2x2ZXMgYm9uZHMgYmV0d2VlbiBtdWx0aXBsZSwgZm9sZGVkIHBvbHlwZXB0aWRlIGNoYWlucy4=[Qq]
[c]IGo=[Qq]
[f]IE5vIExldHRlciAmIzgyMjA7SiYjODIyMTsgaXMgc2hvd2luZyB0aGUgdGhpcmQgbGV2ZWwgKHRlcnRpYXJ5IGxldmVsKSBvZiBwcm90ZWluIHN0cnVjdHVyZS4gNHRoIGxldmVsIG9yIHF1YXRlcm5hcnkgc3RydWN0dXJlIGludm9sdmVzIGJvbmRzIGJldHdlZW4gbXVsdGlwbGUsIGZvbGRlZCBwb2x5cGVwdGlkZSBjaGFpbnMu[Qq]
[c]IG s=[Qq]
[f]IFdheSB0byBnbyEgTGV0dGVyICYjODIyMDtLJiM4MjIxOyBpcyBzaG93aW5nIDR0aCBsZXZlbCwgb3IgcXVhdGVybmFyeSBzdHJ1Y3R1cmUu[Qq]
[/qwiz]
8. Denaturation can change a protein’s shape and interfere with its function
As we’ve seen above, many of the bonds that determine a protein’s 3-D shape are relatively weak, such as hydrogen bonds. Other interactions, such as ionic bonds, only occur if a solution has a specific pH.
Proteins, in other words, can be delicate, and their function can be affected by changes in their environment. A protein that loses its function because of environmental change —usually heat or pH — is said to be denatured, and the process by which this happens is called denaturation.
This might seem abstract and far from your experience, but it’s not. The white of an egg is a protein called ovalbumin. When a bird lays her egg, the protein is in a liquid form. That’s the egg white that you see when you crack an egg.
When you cook an egg, the cooking changes the ovalbumin protein in a way that creates new chemical bonds between ovalbumin molecules. Instead of being able to move freely, the ovalbumin becomes locked in place. That’s why cooked egg white is a solid.
This change from liquid protein to solid protein is an example of denaturation. You’ve changed the nature of the protein.
 |
 |
| raw egg white | Cooked egg white (denatured) |
Understanding denaturation also gives us insights into how enzymes are affected by changes in their environment. Enzymes are protein catalysts that speed up chemical reactions in living things. The diagram below shown, in cartoon form, how an enzyme like lactase might work to break the disaccharide lactose into two monosaccharides, glucose and galactose
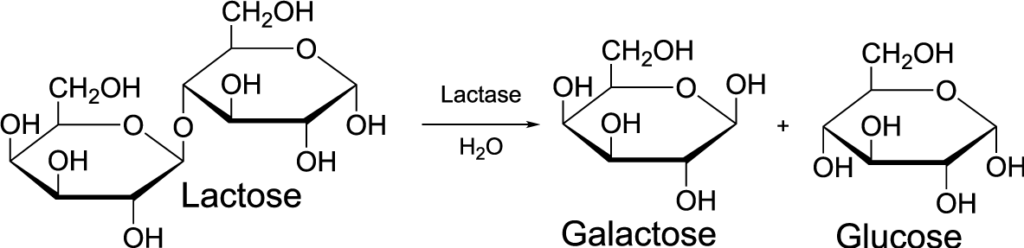
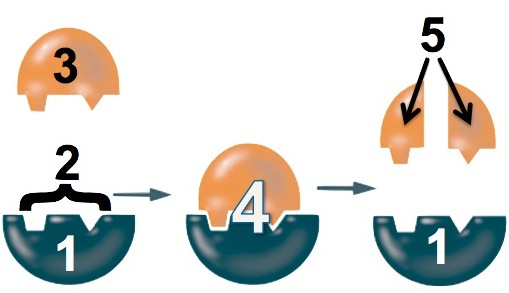
- The lactase enzyme (“1”) binds with the lactose (“3”). The binding is only possible because a specific part of the enzyme, its active site (“2”), has a shape that complements the shape of the lactose.
- While the enzyme is bound to the lactose, the enzyme changes shape to stress the bonds that hold the glucose and galactose monomers together.
- The bond breaks, and the glucose and galactose are released from the enzyme.
While the details of different enzyme-catalyzed reactions will vary, the key point is that the enzyme can catalyze the reaction only if it can fit together with whatever molecule it interacts with. That fit is as specific as the ability of a key to open a lock. Change the key’s shape, and the lock won’t open.
With these ideas in hand, let’s look at the graph below, which shows the relationship between the rate of an enzyme-catalyzed reaction, the percentage of active enzyme, and temperature.
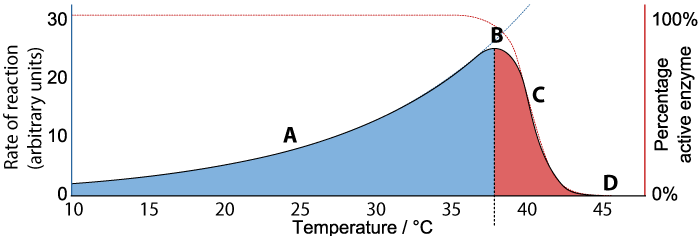
We’ll consider rate first. Follow the black line at “A”, “B” and “C” while looking at the Y axis on the left. As the temperature increases from 10° C to about 37°C (“A”), the rate of the reaction also increases. At about 37°C, the reaction reaches a peak (“B”). As temperature continues to increase much beyond 37°C, the rate of the reaction falls (“C”) until at 45°C the rate of the reaction is at zero.
Now consider “percentage of active enzyme” (the red line, and the Y axis on the right). For any temperature between 10° C to just beyond 37°C, the percentage of active enzyme is 100%. But after point “B,” the percentage drops off (“C”) until it reaches zero (“D”).
What’s happening is denaturation. Up to a certain temperature, the enzyme is able to hold its shape, and interact with its substrate to bring about the reaction. In fact, as temperature increases up to 37°C, the reaction proceeds faster and faster, because higher temperature means more molecular movement, and a greater chance that the enzyme will bump into its substrate at the right orientation to bring the reaction about. Beyond 37°C, the kinetic energy in the system starts to disrupt the bonds that are holding the enzyme in its required three dimensional shape. As this shape starts to change, the enzyme becomes unable to bond with its substrate, and the rate of the reaction (along with the amount of active enzyme) falls to zero.
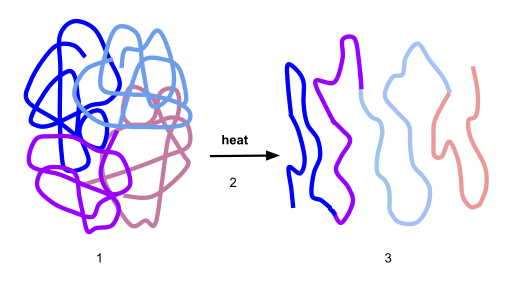 Here’s a visual representation of denaturation. Number “1” shows the protein in its optimal conformation. Heat (“2”) breaks the bond that stabilize this protein’s shape at the secondary and tertiary level. As a result, the enzyme unfurls into a polypeptide.
Here’s a visual representation of denaturation. Number “1” shows the protein in its optimal conformation. Heat (“2”) breaks the bond that stabilize this protein’s shape at the secondary and tertiary level. As a result, the enzyme unfurls into a polypeptide.
Sometimes, like when you cook an egg and change liquid albumin into solid egg white, the denaturation is irreversible. In other words, you might let your egg white cool down, but it’s not going to liquify again. On the other hand, some proteins can temporarily be denatured (usually by changes in pH). When the pH is reset to the original level, the amino acids in the polypeptide might be able to interact in a way that restores the protein’s original shape. That process is called renaturation.
9. Proteins Cumulative Quiz
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Proteins cumulative quiz (HS)”]
[h]Proteins Cumulative Quiz
[i]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14b0ca141e1d0e” question_number=”1″] In the diagram below, which number shows a ribosome synthesizing a protein?
[textentry single_char=”true”]
[c]ID c=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzcmIzgyMjE7IHNob3dzIGEgcmlib3NvbWUgdHJhbnNsYXRpbmcgbVJOQSBpbnRvIGEgcG9seXBlcHRpZGUu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgcmlib3NvbWUgaXMgaW4gdGhlIGN5dG9wbGFzbSAodGhlIHJlZ2lvbiBkZXNpZ25hdGVkIGJ5IG51bWJlciAmIzgyMjA7Ni4mIzgyMjE7[Qq]
[c]IEVudGVyIGxldHRlcg==[Qq]
[f]IE5vLg==[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14b0868ec5490e” question_number=”2″] In the diagram below, which number shows a newly synthesized protein?
[textentry single_char=”true”]
[c]ID g=[Qq]
[f]IE5pY2Ugam9iLiAmIzgyMjA7OCYjODIyMTsgc2hvd3MgYSByaWJvc29tZSB0cmFuc2xhdGluZyBtUk5BIGludG8gYSBwb2x5cGVwdGlkZSwgYW5kIHRoYXQgcG9seXBlcHRpZGUgY2hhaW4gaXMgaW5kaWNhdGVkIGJ5IHRoZSBudW1iZXIgJiM4MjIwOzguJiM4MjIxOw==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgcmlib3NvbWUgYXQgbnVtYmVyICYjODIyMDs3LiYjODIyMTsgV2hhdCBpcyBpdCBzeW50aGVzaXppbmc/[Qq]
[c]IEVudGVyIGxldHRlcg==[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14b043096c750e” question_number=”3″] In the diagram below, which number indicates the polypeptide backbone?
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IEdvb2Qgd29yayAuICYjODIyMDsxJiM4MjIxOyBpbmRpY2F0ZXMgdGhlIHBvbHlwZXB0aWRlIGJhY2tib25lIChldmVyeXRoaW5nIGJ1dCB0aGUgc2lkZSBjaGFpbnMu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgcG9seXBlcHRpZGUgYmFja2JvbmUgaXMgZXZlcnl0aGluZyBidXQgdGhlIHNpZGUgY2hhaW5zIChhbHNvIGtub3duIGFzIFIgZ3JvdXBzKS4=[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14aedece51310e” question_number=”8″] In the diagram below, a hydrogen bond is shown at
[textentry single_char=”true”]
[c]ID U=[Qq]
[f]IEV4Y2VsbGVudCEgQSBoeWRyb2dlbiBib25kIGlzIHNob3duIGF0ICYjODIyMDs1LiYjODIyMTs=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGlzIGRpYWdyYW0gaXMgYWJvdXQgc2Vjb25kYXJ5IHN0cnVjdHVyZS4gSW4gdGhlIGRpYWdyYW0gYWJvdmUsIHdoaWNoIGJvbmQgc2VlbXMgbGlrZSBpdCB3b3VsZCBwbGF5IHRoZSByb2xlIG9mIGZvcm1pbmcgYW5kIHN0YWJpbGl6aW5nIHRoZSBjb3Jrc2NyZXcgc2hhcGUgdGhhdCB0aGlzIHNlY3Rpb24gb2YgYSBwcm90ZWluIGhhcyB0d2lzdGVkIGludG8/[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14ad818f59990e” question_number=”13″] In the diagram below, the polypeptide backbone is indicated by number
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IFllcy4gVGhlIHBvbHlwZXB0aWRlIGJhY2tib25lIGlzIGluZGljYXRlZCBieSBudW1iZXIgJiM4MjIwOzEuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIG51bWJlciB0aGF0JiM4MjE3O3Mgbm90IHBvaW50aW5nIHRvIGFueSBvZiB0aGUgaW50ZXJhY3Rpbmcgc2lkZSBjaGFpbnMu[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14ad3bb5f4e10e” question_number=”14″] In the diagram below, a hydrogen bond between two side chains is indicated by number
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IE5pY2UuIE51bWJlciAmIzgyMjA7MiYjODIyMTsgc2hvd3MgdHdvIHNpZGUgY2hhaW5zIHRoYXQgYXJlIGh5ZHJvZ2VuIGJvbmRpbmcgd2l0aCBvbmUgYW5vdGhlciwgY2F1c2luZyBhIGJlbmQgaW4gdGhlIHBvbHlwZXB0aWRlIGJhY2tib25lLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhbiDigJRPSCBncm91cCBhdCB0aGUgZW5kIG9mIGEgc2lkZSBjaGFpbi4gVGhlc2UgZ3JvdXBzIGFyZSBmcmVxdWVudGx5IGludm9sdmVkIGluIGh5ZHJvZ2VuIGJvbmRzLg==[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14ac60d997290e” question_number=”17″] In the diagram below, a tertiary interaction involving ionic bonding is shown at
[textentry single_char=”true”]
[c]ID U=[Qq]
[f]IEdyZWF0ISBOdW1iZXIgJiM4MjIwOzUmIzgyMjE7IHNob3dzIGFuIGlvbmljIGJvbmQgYmV0d2VlbiBzaWRlIGNoYWlucy4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJb25pYyBib25kcyBpbnZvbHZlIHRyYWRpbmcgZWxlY3Ryb25zLCB3aGljaCByZXN1bHRzIGluIGZ1bGx5IGNoYXJnZWQgYXRvbXMgb3IgZnVuY3Rpb25hbCBncm91cHMuIExvb2sgYXQgdGhlIGRpYWdyYW0gYWJvdmUsIGFuZCBmaW5kIHBvc2l0aXZlIGFuZCBuZWdhdGl2ZSBjaGFyZ2VzLCBhbmQgdGhlcmUgeW91JiM4MjE3O2xsIGZpbmQgYW4gaW9uaWMgYm9uZC4=[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14ac1b0032710e” question_number=”18″] In the diagram below, primary structure is indicated by number
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IEV4Y2VsbGVudC4gUHJpbWFyeSBzdHJ1Y3R1cmUgaXMgaW5kaWNhdGVkIGJ5IG51bWJlciAmIzgyMjA7MS4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBQcmltYXJ5IHN0cnVjdHVyZSBpcyB0aGUgc2VxdWVuY2Ugb2YgYW1pbm8gYWNpZHMuIFdoaWNoIG51bWJlciBzaG93cyB0aGF0IHNlcXVlbmNlICh3aXRob3V0IGFueSBvZiB0aGUgZm9sZGluZyBhbmQgYmVuZGluZyB0aGF0IGVtZXJnZXMgYXQgaGlnaGVyIGxldmVscyBvZiBzdHJ1Y3R1cmU/[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14abd77ad99d0e” question_number=”19″] In the diagram below, an alpha helix is indicated by number
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IFRlcnJpZmljLiBBbiBhbHBoYSBoZWxpeCAoYSBzZWNvbmRhcnkgbGV2ZWwgb2YgcHJvdGVpbiBzdHJ1Y3R1cmUpIGlzIHNob3duIGF0IG51bWJlciAmIzgyMjA7Mi4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBTZWNvbmRhcnkgc3RydWN0dXJlIGludm9sdmVzIGh5ZHJvZ2VuIGJvbmRpbmcgYmV0d2VlbiB0aGUgYW1pbm8gYW5kIGNhcmJvbnlsIGZ1bmN0aW9uYWwgZ3JvdXBzIHdpdGhpbiBhIHByb3RlaW4mIzgyMTc7cyBwb2x5cGVwdGlkZSBiYWNrYm9uZS4gVHdvIHByaW1hcnkgc2Vjb25kYXJ5IGxldmVsIHN0cnVjdHVyZXMgYXJlIGFscGhhIGhlbGljZXMgYW5kIGJldGEgcGxlYXRlZCBzaGVldHMuIFRoZSBoZWxpeCBpcyBsaWtlIGEgY29ya3NjcmV3ICh3aGljaCBzaG91bGQgaGVscCB5b3UgZmluZCBpdCku[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14ab8aa551390e” question_number=”20″] In the diagram below, a protein’s tertiary structure is indicated by number
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IE5pY2UuIFRlcnRpYXJ5IHN0cnVjdHVyZSBpcyBpbmRpY2F0ZWQgYnkgbnVtYmVyICYjODIyMDs0LiYjODIyMTs=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUZXJ0aWFyeSBzdHJ1Y3R1cmUgaXMgd2hhdCBhcmlzZXMgYnkgYm9uZGluZyBiZXR3ZWVuIHNpZGUgY2hhaW5zIHdpdGhpbiBhIHNpbmdsZSBwb2x5cGVwdGlkZSBjaGFpbiwgd2hpY2ggcmVzdWx0cyBpbiBhIHZlcnkgY29tcGxleCB0aHJlZSBkaW1lbnNpb25hbCBzaGFwZS4=[Qq]
[q json=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14ab4023d4b90e” question_number=”21″] In the diagram below, a protein’s quaternary structure is indicated by number
[textentry single_char=”true”]
[c]ID U=[Qq]
[f]IFBlcmZlY3QuIFF1YXRlcm5hcnkgc3RydWN0dXJlIGlzIGluZGljYXRlZCBieSBudW1iZXIgJiM4MjIwOzUuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBRdWF0ZXJuYXJ5IHN0cnVjdHVyZSBpcyB3aGF0IGFyaXNlcyBieSBpbnRlcmFjdGlvbnMgYmV0d2VlbiBtdWx0aXBsZSBwb2x5cGVwdGlkZSBjaGFpbnMuIEJvbmRzIGJldHdlZW4gdGhlc2UgY2hhaW5zIGxlYWQgdGhlbSB0byBmb3JtIGEgbXVsdGktcG9seXBlcHRpZGUgY2hhaW4gcHJvdGVpbi4gV2hpY2ggbnVtYmVyIHNob3dzIG11bHRpcGxlIHBvbHlwZXB0aWRlIGNoYWlucz8=[Qq]
[q json=”true” multiple_choice=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14aaeea6348d0e” question_number=”22″] In the graph below, which letter shows where the enzyme has become completely denatured?
[c]IEE=
[c]IEI=[Qq]
[c]IEM=[Qq]
[c]IE Q=[Qq]
[f]IE5vLiBBdCAmIzgyMjA7YSYjODIyMTsgdGhlIGJsdWUgYXJlYSB1bmRlciB0aGUgbGluZSBpbmRpY2F0ZXMgdGhhdCB0aGUgcmVhY3Rpb24gaXMgbm90IG9ubHkgcHJvY2VlZGluZywgYnV0IGFjY2VsZXJhdGluZy4gVGhlIHJlZCBsaW5lIGF0IHRvcCBpbmRpY2F0ZXMgdGhhdCAxMDAlIG9mIHRoZSBlbnp5bWUgaXMgYWN0aXZlLiBXaGVyZSBvbiB0aGlzIGdyYXBoIGhhcyB0aGUgcmVhY3Rpb24gcmF0ZSBmYWxsZW4gdG8gemVybywgYW5kIDAlIG9mIHRoZSBlbnp5bWUgaXMgYWN0aXZlPw==[Qq]
[f]IE5vLiBBdCBCLCB0aGlzIGVuenltZSYjODIxNztzIGFjdGl2aXR5IGlzIGF0IGl0cyBoaWdoZXN0IHJhdGUuIEFsc28sIHRoZSByZWQgbGluZSBhdCB0b3AgaW5kaWNhdGVzIHRoYXQgMTAwJSBvZiB0aGUgZW56eW1lIGlzIGFjdGl2ZS4gV2hlcmUgb24gdGhpcyBncmFwaCBoYXMgdGhlIHJlYWN0aW9uIHJhdGUgZmFsbGVuIHRvIHplcm8sIGFuZCAwJSBvZiB0aGUgZW56eW1lIGlzIGFjdGl2ZT8=[Qq]
[f]IE5vLiBBdCBDLCB0aGlzIGVuenltZSYjODIxNztzIGFjdGl2aXR5IGlzIHN0YXJ0aW5nIHRvIGRlY2xpbmUsIGJ1dCBzb21lIG9mIGl0IGlzIHN0aWxsIGFjdGl2ZS4gV2hlcmUgb24gdGhpcyBncmFwaCBoYXMgdGhlIHJlYWN0aW9uIHJhdGUgZmFsbGVuIHRvIHplcm8sIGFuZCAwJSBvZiB0aGUgZW56eW1lIGlzIGFjdGl2ZT8=[Qq]
[f]IEV4Y2VsbGVudC4gQXQgJiM4MjIwO0QmIzgyMjE7IHRoaXMgZW56eW1lJiM4MjE3O3MgYWN0aXZpdHkgaGFzIGZhbGxlbiB0byB6ZXJvLCBhbmQgMCUgb2YgdGhlIHByb3RlaW4gaXMgYWN0aXZlLg==[Qq]
[q json=”true” multiple_choice=”true” xx=”2″ dataset_id=”SMV_biochem_8_proteins_cumulative|14aa6e97a6910e” question_number=”23″] A denatured protein is shown at
[c]IDE=
[f]IE5vLiAmIzgyMjA7MSYjODIyMTsgaXMgdGhlIGNvcnJlY3RseSBmb2xkZWQgcHJvdGVpbiB3aXRoIGl0cyAzLUQgc3RydWN0dXJlIGludGFjdC4=[Qq]
[c]IDI=[Qq]
[f]IE5vLiAmIzgyMjA7MiYjODIyMTsgaXMgaGVhdCwgd2hpY2ggaXMgY2F1c2luZyB0aGUgZGVuYXR1cmluZy4=[Qq]
[c]ID M=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzMmIzgyMjE7IHJlcHJlc2VudHMgdGhlIGRlbmF0dXJlZCBwcm90ZWluLiBJdCB3YXMgZGVuYXR1cmVkIGJ5IHRoZSBoZWF0IHdoaWNoIGNhdXNlZCB0aGUgYm9uZHMgbWFpbnRhaW5pbmcgaXRzIHNlY29uZGFyeSBhbmQgdGVydGlhcnkgc3RydWN0dXJlIHRvIHVucmF2ZWwu[Qq]
[x][restart][/qwiz]
Links
-
- Nucleic Acids Overview (the last tutorial in this series)
- The Molecules of Life Main Menu
