1. Embryology as Evidence for Evolution
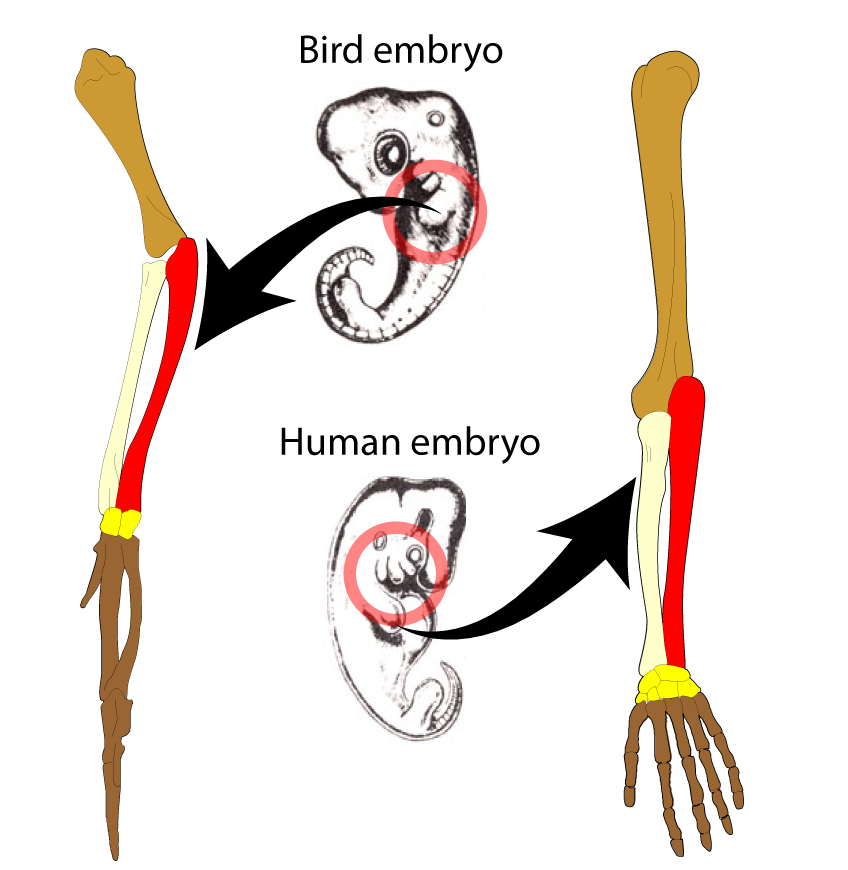 We learned previously that one way that we know that two structures are homologous is because they originate in a similar patch of embryonic tissue. In the diagram to the left, the same patch of tissue develops into the wing of a bird and the arm of a human being. As the animal develops into its adult form, the structure becomes modified to serve a different function.
We learned previously that one way that we know that two structures are homologous is because they originate in a similar patch of embryonic tissue. In the diagram to the left, the same patch of tissue develops into the wing of a bird and the arm of a human being. As the animal develops into its adult form, the structure becomes modified to serve a different function.
The study of embryos reveals additional evidence for evolution. To see how, let’s return to an image from Unit 6 about how developmental genes are expressed in a hierarchical sequence.
As we saw in Unit 6, the images below show embryos from different types of vertebrates — animals with backbones — at equivalent phases of development. The top row consists of photographs with some modifications (a yolk sac removed in the salamander). The bottom row shows drawings by Ernst Haeckel, a famous 19th-century biologist.
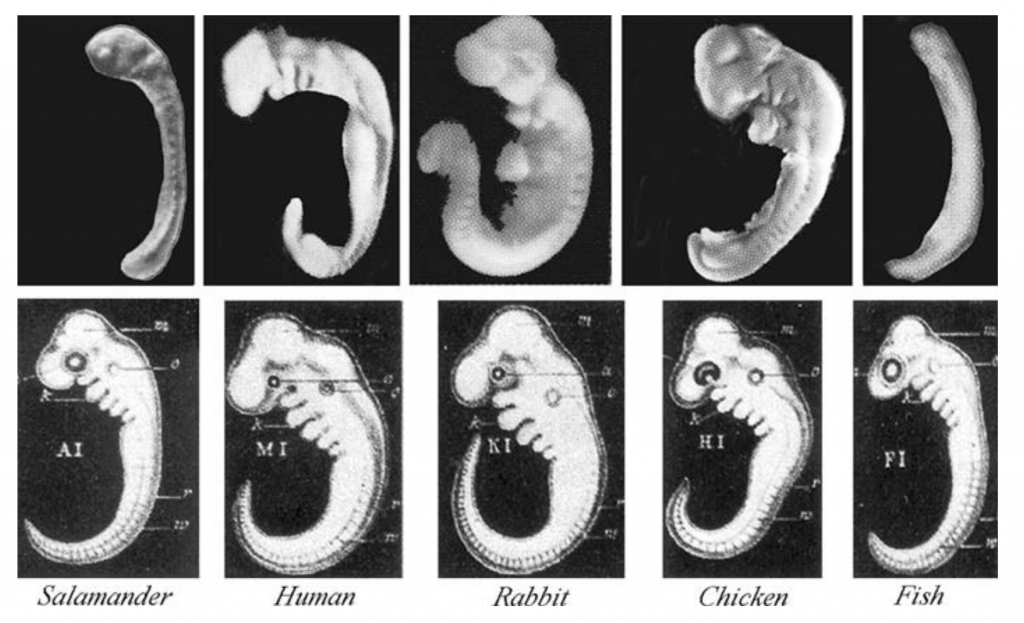
The similarity in embryonic form provides a few additional pieces of evidence for evolution. First, the similarity of form is a homologous trait. The common pattern of development in all vertebrates is a feature that was inherited from a common ancestor.
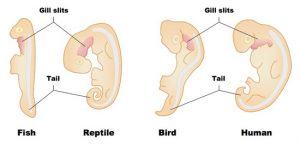
Second, examining embryos reveals two vestigial traits that disappear later in development.
- Pharyngeal slits (also called “gill slits”) appear in humans, rabbits, and chickens. These slits are openings from the neck into the back of the mouth. Fish never lose these slits: they use them to pass water over their gills so that they can absorb oxygen from water. In all the tetrapod vertebrates (the four-limbed amphibians, reptiles, birds, and mammals), these slits disappear later in development.
- Tails appear in humans and chickens and then disappear later in development.

Why do humans, rabbits, and chickens expend the energy to create embryonic gill slits, only to break them down and absorb them later? It’s because having gill slits is part of a developmental pattern inherited from a vertebrate ancestor. In early embryos, the genes for this plan are still expressed.
The same is true of the tail. All vertebrate embryos have a pronounced tail. That includes animals like humans and birds, which as adults don’t have tails. Why do these embryos have tails? Because having a tail is a vestigial trait. It’s a vestige of a developmental pattern inherited from a common ancestor.
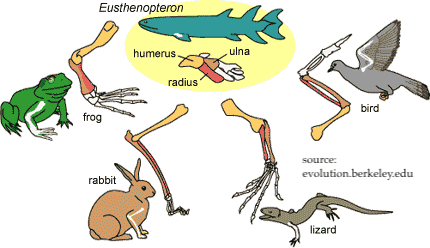 What’s the takeaway? Embryological development allows us to more clearly see the homologies that come about through adaptive radiation.
What’s the takeaway? Embryological development allows us to more clearly see the homologies that come about through adaptive radiation.
- All vertebrates descend from a common ancestor.
- As vertebrates develop, their body plan unfolds in a way common to all vertebrates. That embryonic pattern is itself a homology.
- As development continues, the form of each lineage diverges. The same patch of tissue will develop into a wing of a bird or the forelimb of a lizard, rabbit, or frog.
- The homologous structure of the forelimb in the adult form of each animal reflects adaptive radiation from a common ancestor.
Finally, note that this same pattern of embryonic similarity followed by development into diverse forms can be found in any other group of organisms descended from a common ancestor. You can learn about the differentiation of insect mouthparts in this article on Wikipedia.
2. Flashcards: Embryological Evidence for Evolution
[qdeck bold_text=”false” style=”width: 600px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-Embryological evidence for evolution (v2.0)”]
[h] Embryological evidence for evolution
[q] Why is the similar embryonic form of related animals (such as vertebrates) evidence for evolution?
[a] The similar embryonic form of related animals (such as vertebrates) is evidence for evolution because shows descent from a common ancestor.
[q] How can you explain the appearance of features like gill slits and tails during human development, even though these structures disappear later?
[a] Embryonic gill slits and a tail are vestigial features. Their appearance reflects a pattern of development that we inherited from an ancestor.
[q] What’s the connection between embryological development, adaptive radiation, and homologous structures?
[a] As shown by their homologous structures and common pattern of embryonic development, all vertebrates are descended from a common ancestor. That common ancestor, through adaptive radiation, evolved into different forms.
[x] [restart]
[/qdeck]
3. Molecular Homologies Part 1: Proteins.
Above and in previous tutorials, we’ve discussed how homologous structures serve as evidence for evolution. Molecules can be homologous too, establishing another line of evidence for evolution.
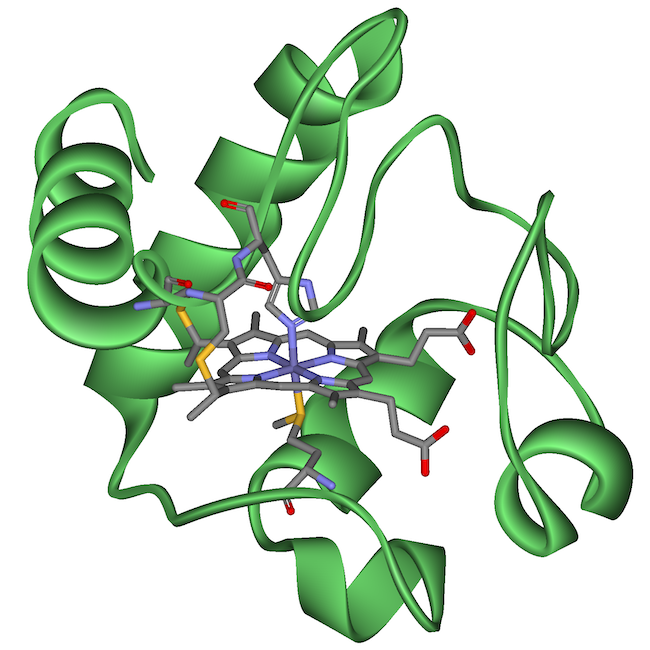
The table below shows two proteins that are widely shared among many species.
 |
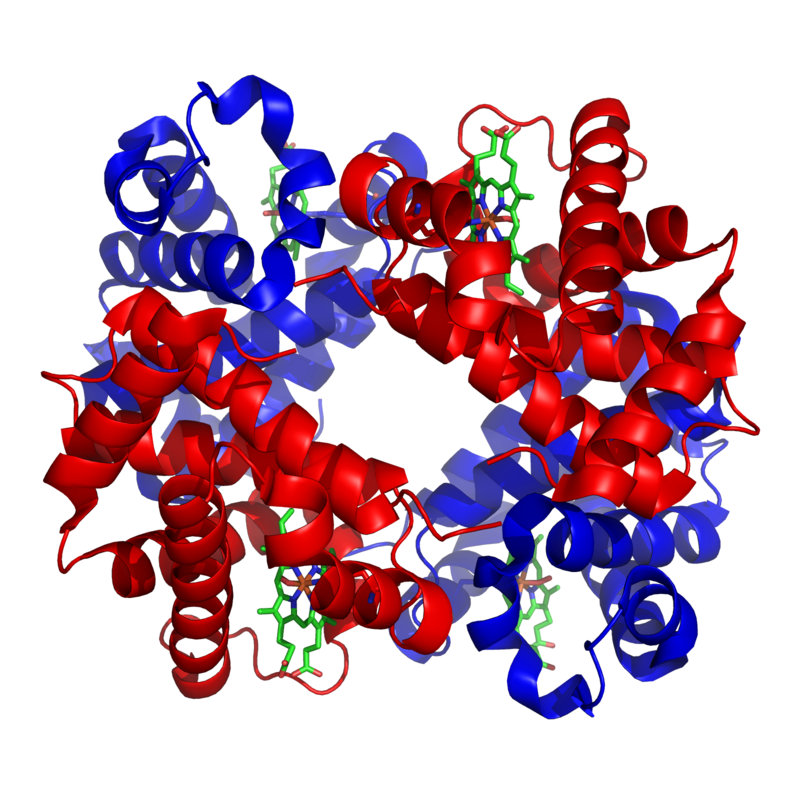 |
| Cytochrome c is a protein that’s used in cellular respiration. | Hemoglobin is a protein used to carry oxygen in red blood cells. |
Cytochrome c and hemoglobin are found in many species with either an identical or highly similar sequence of amino acids and a correspondingly similar three-dimensional shape. Why do so many different species share these molecules? Because they inherited the genes for making these molecules from a common ancestor.
To learn more, complete the following interactive reading.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Interactive Reading: Molecular Homologies and Evolution (v2.0)”]
[h] Interactive Reading: Molecular Homologies and Evolution
[i]
[q labels = “top”]Cytochrome c is a widely shared protein that’s built of 104 linked _________ acids. We’ve previously discussed how the forelimbs of all vertebrates (animals with backbones) are _______________. The same is true of cytochrome c shared by various vertebrate species. Both the forelimb structure in vertebrates and the cytochrome c structures shared by vertebrates (and other species without backbones) were derived from a common ______________ and were _____________ in the descendants.
[l]ancestor
[f*] Excellent!
[fx] No, that’s not correct. Please try again.
[l]amino
[f*] Good!
[fx] No. Please try again.
[l]modified
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l] homologous
[f*] Correct!
[fx] No. Please try again.
[q labels = “top”]The next slide will show a table that shows the number of amino acid differences found between the cytochrome c molecules of human beings and seven other species. In the table, the species names aren’t filled in yet (that’s your job). The more closely __________ a species is to human beings, the fewer _____________ we’d expect to find in the amino acids that make up this protein. Use that hint to complete the table on the next slide.
[l]differences
[f*] Correct!
[fx] No, that’s not correct. Please try again.
[l]related
[f*] Good!
[fx] No. Please try again.
[q labels = “right”]Use logic and trial-and-error to complete the table below. Don’t worry about getting a wrong answer (but remember the hint from the previous slide: the more closely related a species is to human beings, the fewer amino acid differences it will have).
| Organism | Number of amino acid differences from humans |
| ___________________ | 0 |
| ___________________ | 1 |
| ___________________ | 9 |
| Pigeon | 12 |
| ___________________ | 18 |
| ___________________ | 25 |
| Yeast | 40 |
[l]Bullfrog
[f*] Good!
[fx] No. Please try again.
[l]chimpanzee
[f*] Correct!
[fx] No. Please try again.
[l]Fruit fly
[f*] Good!
[fx] No. Please try again.
[l]Rabbit
[f*] Great!
[fx] No. Please try again.
[l]Monkey
[f*] Great!
[fx] No. Please try again.
[q labels = “top”]This cytochrome c data confirms other aspects of biology. For example, of the other species in this list, chimpanzees are, in terms of their anatomy and behavior, the most _____________ to humans. Therefore it makes sense that their amino acid sequence for this key protein would be ____________ to the human sequence. Of the mammals listed, the _________ is more different from humans than is the monkey. Therefore, it makes sense that the rabbit’s cytochrome c amino acid sequence would, from among the mammals in the table, also be the most ____________ from the human sequence.
[l]different
[f*] Good!
[fx] No, that’s not correct. Please try again.
[l]identical
[f*] Excellent!
[fx] No. Please try again.
[l]rabbit
[f*] Excellent!
[fx] No. Please try again.
[l]similar
[f*] Great!
[fx] No. Please try again.
[q labels = “top”]The key idea is that the cytochrome c shared by the organisms below is a ________________ structure. The organisms below share it because they inherited the genes for this molecule from a common ______________. Of the organisms below, our most distant relative is ______________ (which, unlike every other organism on the list, is not an ____________, but a fungus). As our most distant relative, it makes sense that its cytochrome c would also be the most _____________. But despite these differences, we still share a common ancestor with yeast. Evolutionarily speaking, it’s our distant ___________.
[l]ancestor
[fx] No. Please try again.
[f*] Good!
[l]animal
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]cousin
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]different
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l]homologous
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]yeast
[fx] No. Please try again.
[f*] Correct!
[q] Let’s learn a bit more about hemoglobin. This is a protein that transports oxygen in red blood cells. The protein consists of four amino acid chains. Two are 141 amino acids long. The other two are 146 amino acids long.
[q labels = “top”]Complete this list of amino acid differences between one of the hemoglobin chains in humans and the corresponding chain in several other species.
[l]27
[f*] Correct!
[fx] No, that’s not correct. Please try again.
[l]one
[f*] Good!
[fx] No, that’s not correct. Please try again.
[l]45
[f*] Good!
[fx] No. Please try again.
[l]eight
[f*] Good!
[fx] No, that’s not correct. Please try again.
[l]62
[f*] Excellent!
[fx] No. Please try again.
[q labels = “top”]The presence of hemoglobin in all of the animals below is a molecular _______________. All of these animals have hemoglobin because they inherited it from a common ______________. The amount of molecular similarity reflects the amount of similarity in body form and other features. Of the animals below, the one most similar to humans is the ______________. The similarity in body structure is matched by the similarity in the _______________ making up hemoglobin, which only differs by _______. The more _____________ the animal is from a human being, the more differences will be found in the sequence of the _____________ in their red blood cells.
[l]amino acids
[fx] No. Please try again.
[f*] Great!
[l]ancestor
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]different
[fx] No. Please try again.
[f*] Good!
[l]gorilla
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]hemoglobin
[fx] No. Please try again.
[f*] Great!
[l]homology
[fx] No. Please try again.
[f*] Correct!
[l]one
[fx] No, that’s not correct. Please try again.
[f*] Good!
[x][restart]
[/qwiz]
What’s the takeaway? The sharing of common proteins like cytochrome c and hemoglobin among animals isn’t unusual. Most of the proteins found in human beings are also found in other animals. The sharing of these proteins is a homology, indicating descent from a common ancestor.
4. Molecular Homologies Part 2: Homologous DNA Sequences and Genes
What’s true of proteins is also true of DNA. The image below shows a stretch of DNA from seven animals. Why is there so much overlap between the sequences? Because all of these animals share a common ancestor. The more recent the common ancestor, the more overlap between the sequences. This similarity in the sequence is called sequence homology, and it’s become the best evidence for drawing evolutionary trees (a topic we’ll take on later in this unit).

Protein-coding genes can also be homologous. Based on how the term “gene” is defined, humans have between 20,000 and 25,000 genes. Almost all of these genes are also found in other animals. We share these genes with other animals because we inherited them from a common ancestor.
Here are two examples:

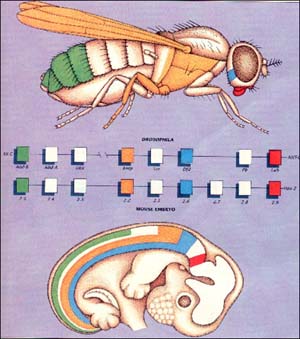
1. Genes coding for eye development
A gene called eyeless codes for eye formation in fruit flies (and other insects). In the image on the right, you can see what the eyeless phenotype looks like.
The eyeless gene is homologous to a gene called small eye in mice (and other mammals).
How do we know that the genes are homologous? First, because of the shared sequence of nucleotide bases in each gene. Second, because of a series of experiments in which these genes were transferred from fruit flies to mice, and vice versa. The eyeless gene has been experimentally inserted into mutant mice embryos that are from a strain of mice that don’t develop eyes. The result? The functional eyeless gene from fruit flies induced eye development in mice (a mammal) The reverse is also true. Inserting the mouse gene into embryos of mutant fruit flies that don’t develop eyes, and eyes will develop.
2. Genes coding for body segments: Almost all animals are built using a similar developmental plan. The plan involves the development of a head, a middle region, and a hind region. In many animals, these regions can be subdivided into segments.
The genes that control how each segment develops are called Hox genes, and they’re shared between insects and mammals. How do we know they’re homologous? Because of sequence homologies, and because of gene transfer experiments. In other words, the same gene that directs the formation of structures on the head in an insect also directs the formation of a head in mammals (including you). These genes are shown in red in the diagram to the right. The same is true of the structures on the rest of the body, from head to tail (even if your tail is just a vestige).
What do these genetic homologies tell us? That mice and fruit flies share a common ancestor from which they both inherited the same genes for eye development and body segment identity. That common ancestor existed 500 million years ago when the line leading to vertebrates split apart from the line leading to arthropods (insects, crabs, spiders, etc.) And just to make this personal, that means that insects are our very ancient cousins. Think of that the next time you swat a fly.
5. Genes can be vestigial, too: pseudogenes
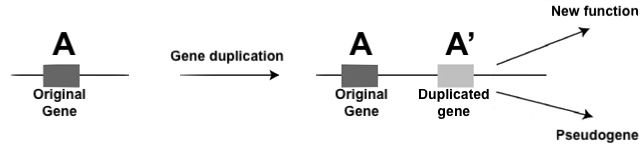
If we think about vestigial structures like the hind limbs of whales, it’s clear that evolution doesn’t necessarily delete the genes for traits that are no longer needed. Rather, the evolutionary process involves diminishing the size of structures (such as the hind limbs of whales) or allowing mutations to accumulate in these structures (such as the vestigial eyes of blind cavefish) so that they’re no longer functional. As Jerry Coyne argues in his book Why Evolution is True, this allows us to make a prediction. In the genomes of many species, we should be able to find “silenced or dead genes: genes that once were useful but are no longer intact or expressed.”
The non-functional remnant of a formerly functional gene is called a pseudogene. Pseudogenes can be silenced for a variety of reasons. The image below shows all of these compressed into one (but note that any one of the causes shown could be sufficient for preventing gene expression).
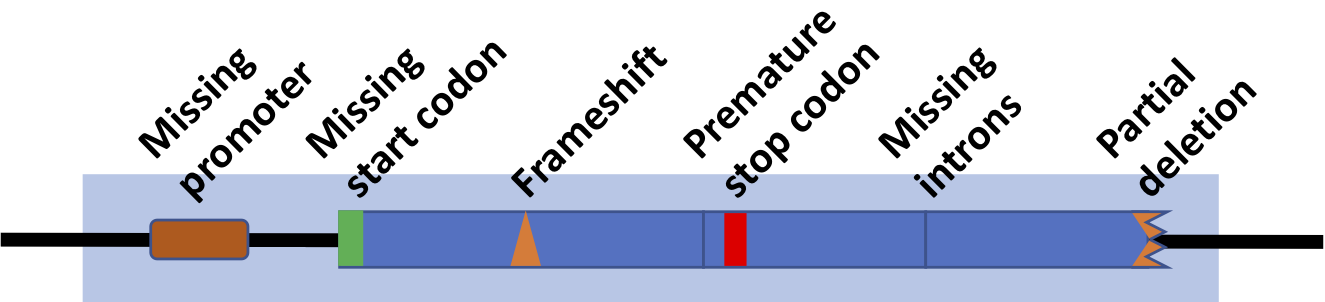
In some cases, pseudogenes result from gene duplication, followed by a mutation in the duplicated gene. In that case, there’s no loss of function, because the original gene is still expressed.
But in other cases, a functional gene becomes silent. An example is the ψGLO pseudogene (the symbol ψ designates a gene as a pseudogene). The functional GLO gene, in other species, codes for L-Gulonolactone Oxidase, an enzyme that’s part of a pathway that converts glucose (C6H12O6) into vitamin C (C6H8O6). L-Gulonolactone Oxidase is found in a functional form in almost all mammals, except for bats, guinea pigs, and primates (the order of mammals that includes monkeys, apes, and humans). The groups that don’t make vitamin C have lost the ability to do so because they live in environments where they could obtain vitamin C from fruit in their environment. Consequently, any mutation that silenced the gene for producing L-Gulonolactone Oxidase would have no negative effect on fitness.
What happens without vitamin C? It took an unintended experiment — sailors undertaking long sea voyages without any available fruit — to see that vitamin C deficiency causes scurvy, the symptoms of which include anemia, weakness, spontaneous bleeding (especially in the gums), loss of teeth, pain in the limbs, and swelling. Prolonged vitamin C deficiency can be fatal. In 1757, a Scottish physician named James Lind persuaded the British Navy to start issuing lime juice to sailors. That ended scurvy in the British Navy, and is why British sailors (and even British people) are sometimes called “Limeys.”
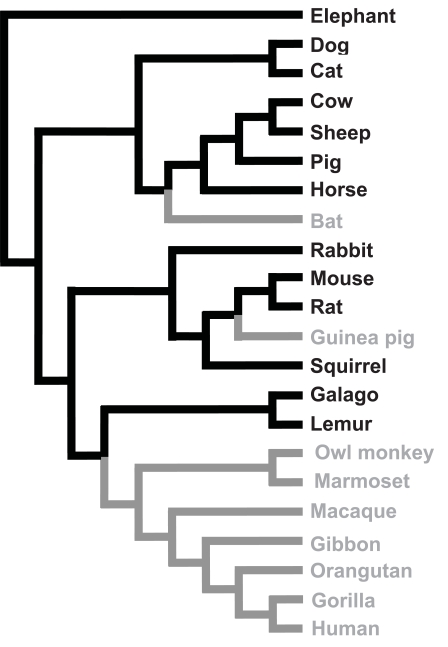
About 40 million years ago, an ancient early primate developed a mutation that deactivated the GLO gene. This created the ψGLO pseudogene, which was passed on to all of his or her descendants, which now includes almost all the primates, except for lemurs and Galagos.
In a few other mammal lineages — guinea pigs and bats — the GLO gene was also silenced, and these lineages have their own version of the ψGLO pseudogene. But because this silencing happened independently from the silencing that happened in primates, the bat and guinea pig pseudogenes are significantly different from the one shared by primates (source: Coyne, 2009). In other words, the existence of the ψGLO pseudogene in primates, bats, and guinea pigs is another example of parallel evolution.
As with shared genes and amino acid sequences for cytochrome c and hemoglobin, the ψGLO pseudogene is not in any way remarkable or unusual. Pseudogenes are common in the human genome. We might have as many pseudogenes as we have genes. In the two decades since the human genome was first sequenced, the number of pseudogenes that have been identified has come to number in the thousands (compared to the 20,000 or so protein-coding genes in the human genome).
6. Flashcards: Molecular Evidence for Evolution
[qdeck bold_text=”false” style=”width: 600px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-Molecular evidence for evolution (v2.0)”]
[h]Molecular evidence for evolution
[q]Shared molecules such as cytochrome c and hemoglobin are evidence for evolution because …
[a]Shared molecules such as cytochrome c and hemoglobin are evidence for evolution because these molecules are evidence of descent with modification. The molecules’ shared structure and sequences can only be explained as homologies, and they indicate that the species that share these proteins must have had a common ancestor.
[q]Cytochrome c is a protein used in _____________ ________________. It’s widely shared among eukaryotic species and is found in [try to list a few other species that also use cytochrome c]
[a]Cytochrome c is a protein used in cellular respiration. It’s widely shared among eukaryotic species and is found in such species as humans, other mammals, insects, and even yeast.
[q]Proteins that are shared among species show a predictable pattern. The more closely the species are related…
[a]Molecules that are shared among species show a predictable pattern. The more closely the species are related the more the sequence of amino acids in those proteins will overlap.
[q]How does the information in the image below serve as evidence of evolution?
[a]The information in the diagram below provides two pieces of evidence for evolution. First, the fact that humans and animals share hemoglobin is evidence of homology on a molecular level. We share hemoglobin with these species because we share a common ancestor. Second, the degree to which our hemoglobin is similar matches and confirms homologies that are based on structure.
[q]Of the animals shown below, the greatest difference in hemoglobin is between humans and __________. That indicates that…
[a]Of the animals shown below, the greatest difference in hemoglobin is between humans and frogs. That indicates that humans and frogs are the least closely related, and share the most distant common ancestor.
[q]We share most of the 20,000 or so genes in the human genome with other animals. That’s evidence that …
[a]We share most of the 20,000 or so genes in the human genome with other animals. That’s evidence that we share a common ancestor with other animals.
[q]What is a sequence homology? How do sequence homologies serve as evidence for evolution?
[a]A sequence homology is a shared sequence of nucleotides or amino acids found in different organisms. The shared sequence indicates that the organisms in question share a common ancestor. The more closely aligned the sequenced, the more recent the common ancestor.
[q]How do we know that the eyeless gene in insects and the small eye gene in mammals are homologous? What do these genes say about the relationship between insects and mammals?
[a]We know that the eyeless gene in insects and the small eye gene in mammals are homologous for two reasons. First, because of the sequence homology between these two genes. Second, because of transplantation experiments in which functional versions of each gene can activate eye development in eyeless mutants from the other group.
This homology indicates that mammals and insects share a common ancestor.
[q]Explain the diagram below.
[a]The diagram shows how the genes that control which structures grow on which body segment — Hox genes — are homologous between insects and mammals. That homology points to a common ancestor that mammals share with insects.
[q]What are pseudogenes?
[a]Pseudogenes are genes that have lost their function but persist as recognizable genes in a species’ genome.
[q]How are pseudogenes evidence for evolution?
[a]Pseudogenes are similar to vestigial traits. They can only be explained through their descent with modification from a pre-existing functional gene which either became duplicated or whose function became superfluous, as is the case of the GLO pseudogene in most primates, bats, and guinea pigs.
[q]Based on the evolutionary tree below, tell the story of the evolution of the ψGLO pseudogene
[a]
Based on the evolutionary tree at left, the GLO gene evolved into the ψGLO pseudogene in a species that was ancestral to most (but not all) of the primates. Separate mutations led the GLO gene to evolve into a ψGLO pseudogene in guinea pigs and bats, making this an instance of parallel evolution in these three groups of mammals.
[q]What are some of the defects that can convert a gene into a pseudogene? Try to list at least three.
[a]Any of the defects below can convert a gene into a pseudogene.
[x][restart]
[/qdeck]
7. Checking Understanding Quiz: Embryonic and Molecular Evidence for Evolution
[qwiz style=”width: 600px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-Embryonic and Molecular Evidence for Evolution (v2.0)”]
[h]Embryonic and Molecular Evidence for Evolution
[i]
[q] The embryos of vertebrates such as fish, birds, and mammals develop in similar ways because all of these animals share a common [hangman].
[c]IGFuY2VzdG9y[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] Embryos of humans show various [hangman] traits such as a tail or gill slits that are not present in the adult form.
[c]IHZlc3RpZ2lhbA==[Qq]
[q] The common developmental pattern shown by these vertebrate embryos is evidence of [hangman] with [hangman] from a common ancestor.
[c]IGRlc2NlbnQ=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IG1vZGlmaWNhdGlvbg==[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] Hemoglobin and cytochrome c are both examples of [hangman] at the molecular level.
[c]IGhvbW9sb2dpZXM=[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] Based on molecular data like that shown in the table below, one can infer how closely [hangman] two species are.
[c]IHJlbGF0ZWQ=[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] The more similar the amino acid [hangman] in a protein shared by two species, the more closely related they are.
[c]IHNlcXVlbmNl[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q multiple_choice=”true”] Which of the following statements about human genes is most correct?
[c]IEh1bWFucyBhcmUgdW5pcXVlIGJlY2F1c2UgbW9zdCBvZiBvdXIgZ2VuZXMgYXJlIHVuaXF1ZSB3aXRoaW4gdGhlIGFuaW1hbCBraW5nZG9t[Qq]
[f]IE5vLiBPZiB0aGUgMjAsMDAwICYjODIxMTsgMjUsMDAwIGdlbmVzIGluIHRoZSBodW1hbiBnZW5vbWUsIG9ubHkgYSBmZXcgZG96ZW4gYXJlIHVuaXF1ZSB0byBvdXIgc3BlY2llcy4=[Qq]
[c]IEFsbW9zdCBhbGwgaHVtYW4gZ2VuZXMgaGF2ZSBhIGhvbW9s b2dvdXMgY291bnRlcnBhcnQgaW4gb3RoZXIgYW5pbWFscw==[Qq]
[f]IFllcy4gT2YgdGhlIDIwLDAwMCAmIzgyMTE7IDI1LDAwMCBnZW5lcyBpbiB0aGUgaHVtYW4gZ2Vub21lLCBvbmx5IGEgZmV3IGRvemVuIGFyZSB1bmlxdWUgdG8gb3VyIHNwZWNpZXMuIEFsbCB0aGUgb3RoZXJzIGhhdmUgaG9tb2xvZ291cyBjb3VudGVycGFydHMgaW4gb3RoZXIgc3BlY2llcy4=[Qq]
[c]IEFib3V0IGhhbGYgb2YgaHVtYW4gZ2VuZXMgYXJlIHVuaXF1ZSB0byBodW1hbiBiZWluZ3Mu[Qq]
[f]IE5vLsKgT2YgdGhlIDIwLDAwMCAmIzgyMTE7IDI1LDAwMCBnZW5lcyBpbiB0aGUgaHVtYW4gZ2Vub21lLCBvbmx5IGEgZmV3IGRvemVuIGFyZSB1bmlxdWUgdG8gb3VyIHNwZWNpZXMu[Qq]
[q] Genes or proteins that are widely shared among a variety of species are said to be highly conserved. These genes or proteins are molecular [hangman] and indicate common [hangman].
[c]IGhvbW9sb2dpZXM=[Qq]
[c]IGFuY2VzdHJ5[Qq]
[q] The Hox genes (shown below) control which kind of appendage grows on which body [hangman].
[c]IHNlZ21lbnQ=[Qq]
[q] The similar organization and sequence similarity in the hox genes indicate that these genes are [hangman] between insects and mammals.
[c]IGhvbW9sb2dvdXM=[Qq]
[q]The fact that both human and arthropod development is controlled by hox genes indicates that back in time, these animals share a [hangman] [hangman].
[c]IGNvbW1vbg==[Qq]
[c]IGFuY2VzdG9y[Qq]
[q]Genes that have lost their function are called [hangman]. They can be thought of as a genetic version of a [hangman] trait.
[c]cHNldWRvZ2VuZQ==[Qq]
[c]dmVzdGlnaWFs[Qq]
[q]Pseudogenes can evolve through gene [hangman], or when the [hangman] of a protein-coding gene becomes superfluous.
[c]ZHVwbGljYXRpb24=[Qq]
[c]ZnVuY3Rpb24=[Qq]
[q]The evolution of the ψGLO pseudogene in primates, bats, and guinea pigs is an example of [hangman] evolution.
[c]cGFyYWxsZWw=[Qq]
[x][restart]
[/qwiz]
8. The Deepest Homologies
8a. Homologous features found in all eukaryotes

Our domain, the eukaryotes, arose about 1.5 billion years ago. Despite their incredible diversity, eukaryotes share a single common ancestor. How do we know? Because of the homologous features possessed by all eukaryotes.
Here’s a short list of the homologous features that all eukaryotes share.
- Mitochondria
- Complex cells with internal compartments such as the rough and smooth endoplasmic reticulum, the Golgi apparatus, and lysosomes.
- A membrane-bound nucleus encloses the cell’s chromosomes, separating the chromosomes from the cytoplasm.
- Linear chromosomes (as opposed to the circular chromosomes in bacteria and archaea).
Note that multicellularity is not a shared eukaryotic feature (because many eukaryotes are unicellular).
8b. Homologous features found in all living things
We can go one level deeper, from traits that unify eukaryotes to traits found in all living things.
All living things possess molecular and genetic homologies that point to one of the most profound insights from the study of biology: that all living things are related. We’re all cousins. “We,” in this context, doesn’t just mean all human beings (though that’s true). We’re also cousins to sparrows, leaf-cutter ants, oyster mushrooms, rhododendrons, kelp, Archaeans that live in hot springs, the photosynthetic bacteria that live in the oceans, and the E. coli bacteria that live by the billions in our guts.
To see what these homologies are, and to distinguish the conserved features of all life from the conserved features that bind together the eukaryotes, complete the quiz below.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-We’re all cousins (v2.0)”]
[h]We’re all cousins
[i]
[q]One conserved homology is seen in the molecule that defines life: the genetic material [hangman]
[c]RE5B[Qq]
[q]All living things follow the central dogma: [hangman] makes [hangman] makes [hangman].
[c]RE5B[Qq]
[c]Uk5B[Qq]
[c]cHJvdGVpbg==[Qq]
[q]All living things synthesize their proteins using [hangman], shown at “2” below.
[c]cmlib3NvbWVz[Qq]
[q]All living things are composed of one or more [hangman].
[c]Y2VsbHM=[Qq]
[q]All living things use [hangman] to power cellular work.
[c]QVRQ[Qq]
[q]This one’s tricky. All living things are made of cells. To be a cell, you have to have a selectively permeable [hangman] surrounding the cytoplasm. But that membrane is not, universally speaking, a [hangman] bilayer (see the diagram at the bottom left). That’s because the organisms in an entire domain of life — Archaea — have a different lipid making up their membrane (see the top molecule below: it has branched side chains, and the glycerol has a different orientation)
[c]bWVtYnJhbmU=[Qq]
[c]cGhvc3Bob2xpcGlk[Qq]
[q]All living things translate their RNA into protein using the same [hangman] [hangman]
[c]Z2VuZXRpYw==[Qq]
[c]Y29kZQ==[Qq]
[q]While all living things have genes that get translated into protein, only the eukaryotes have genes that contain [hangman], intervening sequences that need to be edited out.
[c]aW50cm9ucw==[Qq]
[q]If you had to choose one organelle that defines the eukaryotes, it would probably be the [hangman], because when the bacterial ancestor of these organelles was taken up by an archaeal cell (another type of prokaryote), that’s when the eukaryotes were born.
[c]bWl0b2Nob25kcmlh[Qq]
[q]As opposed to the circular chromosomes in prokaryotes, all eukaryotes have [hangman] chromosomes.
[c]bGluZWFy[Qq]
[q]A conserved homology in all eukaryotes is the presence of a(n) [hangman] system, which includes organelles such as the endoplasmic reticulum and the [hangman] complex.
[c]ZW5kb21lbWJyYW5l[Qq]
[c]Z29sZ2k=[Qq]
[q]In all eukaryotes, the chromosomes are enclosed in a [hangman] that separates the chromosomes from the [hangman].
[c]bnVjbGV1cw==[Qq]
[c]Y3l0b3BsYXNt[Qq]
[x]
[restart]
[/qwiz]
9. Flashcards: The Deepest Homologies
[qdeck bold_text=”false” style=”width: 600px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-The Deepest Homologies (v2.0)”]
[h]The Deepest Homologies
[q] What are four homologous features shared by all eukaryotes indicating that they share a common ancestor?
[a] Four homologous features shared by all eukaryotes indicating common ancestry include
- Cells with mitochondria
- Complex cells with internal compartments like the E.R. Golgi, and lysosomes.
- A membrane-bound nucleus separates chromosomes from the cytoplasm.
- Linear chromosomes.
[q] Homologous features shared by all living things, and which indicate that all living things descend from a common ancestor include
[a] Six homologous features shared by all living things, and which indicate that all living things descend from a common ancestor include
- DNA as the genetic material
- A system where DNA is transcribed into RNA, which is translated into protein.
- Using ribosomes to synthesize protein.
- Being composed of cells
- Using ATP to power cellular work.
- Using the same genetic code
[/qdeck]
10. What’s next?
To finish Topics 7.6 – 7.8, Evidence for Evolution, click the following link for Evidence for Evolution cumulative flashcards and a multiple-choice quiz.
