1. Genetic drift can explain allele frequencies in some populations
One principle of population genetics is that allele frequencies in large populations tend to be stable (or change very slowly). In this tutorial, we’ll see how a small population size can lead allele frequencies within a population to randomly change. This change is called genetic drift.
Genetic drift is a change in allele frequencies caused by random selection and reproduction of alleles. In large populations, these random changes are usually averaged out by other random changes in other individuals within the population, so the result is no net effect. Consequently, the effects of genetic drift are usually seen only in populations that are small, or in populations that were very small at some point in their history. The effect of genetic drift is to reduce genetic variation by eliminating alleles from a population’s gene pool. In the most extreme cases, variable alleles can become fixed alleles.
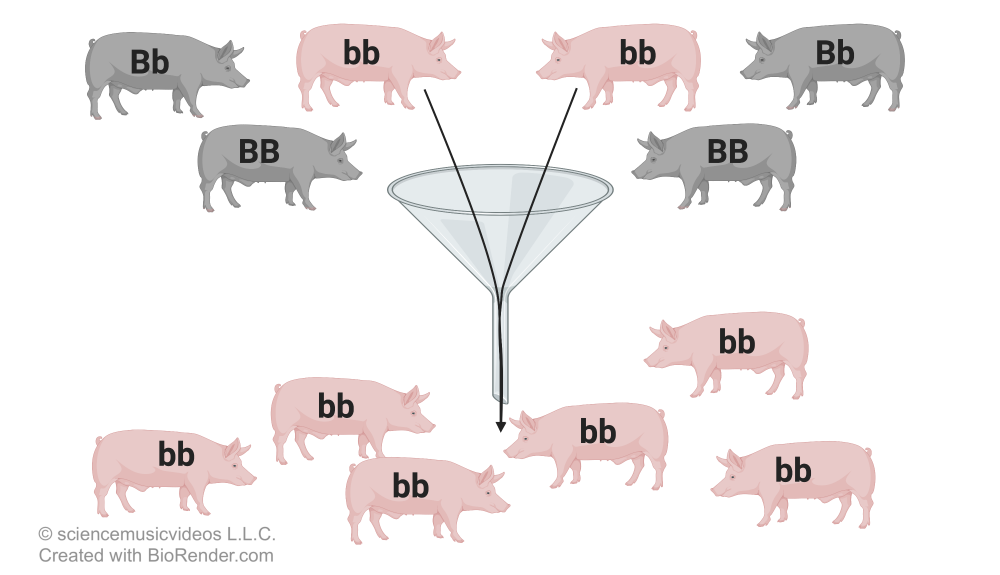
Let’s look again at a population of twenty mice, and examine just one gene. As in previous examples in these tutorials, “A” codes for normal color, and “a” codes for albino coloration.
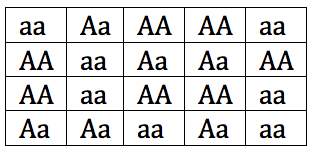
You can see that in this gene pool, the two alleles are evenly distributed. Both “A” and “a” have a frequency of 0.5. Now imagine that a rock slide kills every mouse except for the two on the bottom far right. I’ve put those two in bold, and put a line through all of the other mice.
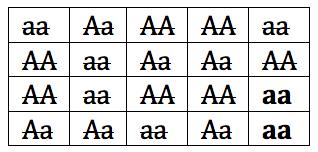
If these two survivors breed, then the new population will have only one allele, “a.” In a few generations, the gene pool will look like this:
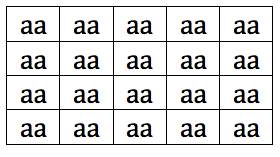
The frequency of “a” is now 1.0, and the frequency of “A” is 0.0. Note that this is NOT about natural selection. There was no survival advantage provided by “a.” The change was completely random. It’s as if we were on a raft that could only hold two individuals. We drifted over to the far corner, and, by chance, picked up two individuals with the genotype “aa.”
Keeping that image in mind, it’s easier to see why genetic drift involves small populations. If our raft were bigger, our random pick up would have resulted in our picking up an “A” allele. The larger the “raft,” the more the sample that we picked up would represent the larger gene pool. Read this description of genetic drift from Wikipedia: the language should now make sense. Click here if you want to read the entire article.
Genetic drift or allelic drift is the change in the frequency of a gene variant (allele) in a population due to random sampling. …The law of large numbers predicts little change over time due to genetic drift when the population is large. When the reproductive population is small, however, the effects of sampling error can alter the allele frequencies significantly. Genetic drift is therefore considered to be a consequential mechanism of evolutionary change primarily within small, isolated populations.
2. Three case studies showing how genetic drift can unfold.
2a. The Amish: Genetic Drift (and loss of diversity) through the Founder Effect

Among the Amish in Eastern Pennsylvania, the frequency of Ellis-van Creveld syndrome is many times higher than it is in the surrounding non-Amish Pennsylvania.
Ellis-van Creveld is a recessive condition whose symptoms include dwarfism, extra fingers and toes (polydactyly), abnormalities of nails and teeth, and a hole between the upper two chambers of the heart. Why would this condition be in such high frequency in this population?
The Pennsylvania Amish were founded by about 200 German immigrants in the mid-1700s. The allele for Ellis-van Creveld syndrome has been traced to one couple from the founding population. And because the Amish tend to marry only within their own culture, the chance of two Amish being heterozygous is much higher than it would be if they married with the surrounding non-Amish population.
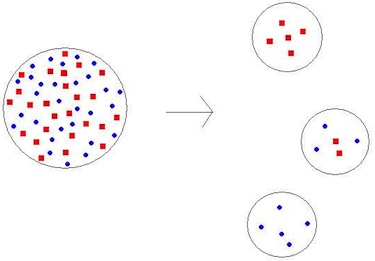
This reduced genetic diversity that results when a population is descended from a small number of colonizing ancestors is known as the “founder effect.” You can see this in the diagram at right. The large circle on the left represents the original population. Red squares and blue diamonds represent the two alleles in this population. Three cases of founding events are shown. In the first (top right) the red allele has become fixed in the population. In the second (right middle) both alleles have been preserved. In the third (bottom right) the blue allele has become fixed in the population. To learn more about the Amish from PBS’s Evolution Library, click here.
2b. Elephant Seals: Genetic Drift and Loss of Diversity Caused by a Recent Population Bottleneck
A population bottleneck has the same result as the founder effect (loss of genetic diversity), but the historical cause is different. In a population bottleneck, a large population is severely reduced. Since the survivors represent only a small sample of the previous population, you can predict that as the population recovers, it will have far less genetic diversity than it did before the bottleneck.

The northern elephant seal, which lives along the West Coast of North America from Alaska down to Baja California, was once hunted to near extinction. By the end of the 1800s, its population may have been as low as 20 individuals. While the population has since rebounded to over 100,000 individuals, genetic studies show that the genetic diversity of the Northern Elephant seal is much less than that of the closely related Southern elephant seal, which lives south of the equator. While the Northern Elephant seals appear to be doing well, biologists fear that their lack of genetic diversity could make them less adaptable to future challenges, including climate change, pollution, and disease.
2.c. Cheetahs: Genetic Drift and Loss of Diversity Caused by an Ancient Population Bottleneck

Whereas the population bottleneck that Northern Elephant Seals suffered was observed in recent history, we can infer that cheetahs suffered a similar bottleneck at some time in the past. The evidence is in the cheetah’s gene pool.
A study published in 1987 compared genetic diversity in cheetahs to genetic diversity in other cat species, and in three non-cat species (click here for an article in the NY Times about this study. Click here for a more in-depth article from Scientific American. For the original study, click here).
In the bar graph below, Cheetahs are represented by the two sets of bars on the far right. Don’t be intimidated by the terms on the axes. “Frequency of polymorphic loci” means how often a specific gene has more than one allele. In other words, for each gene studied, how often is that gene not fixed, but, rather, accompanied by another allele. “Average heterozygosity” looks at how often, for the alleles being studied, individuals are heterozygous.
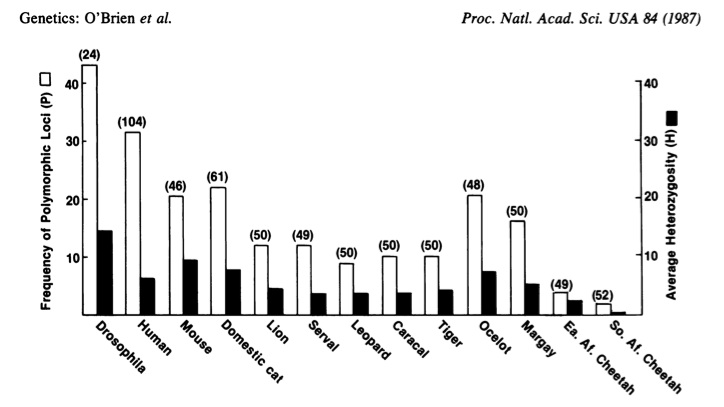
As you can see, the white bars are lower in cheetahs than in any other species. That means that cheetahs have many fewer genes with more than one allele (or, as the graph indicates, many fewer polymorphic loci). Another way of saying that is that many more of the cheetah’s genes are fixed. The black bars are also the lowest, indicating that for any gene studied, cheetahs tend to be much more homozygous, and much less heterozygous.
Simply put, cheetahs have very little genetic diversity. Cheetahs are homozygous for most of their genes. Analysis of 50 blood proteins revealed that the amino acid sequences of those proteins were identical. The genes that code for key tissue protein markers are so similar that cheetahs won’t reject skin grafts from other cheetahs.
Is this a problem? Absolutely. Even with the best of genes, cheetahs would be at risk for extinction from the same loss of habitat that is threatening many African wild species. But if you add to that the cheetah’s low genetic diversity, it puts their population particularly at risk. To begin with, the cheetah’s low variability seems to be causing low fertility. Analysis of cheetah sperm (collected for captive breeding programs) showed that 70% of Cheetah sperm were abnormal. (Original citation from Bioscience, Volume 36, number 6). In captive breeding programs that happen at zoos and wildlife parks, only 10 to 15% of captive cheetahs have been able to successfully breed with one another, and 29.1% of the cubs born die within a year. According to Dr. Stephen O’Brien, head of the research team that produced many of these findings, ”It is not a trivial thing to lose your genetic variation, Genetic variation exists so ecological pressures can be adapted to.”
3. Checking Understanding
This module focused on genetic drift, the founder effect, and the bottleneck effect.
If you feel that you understand this material, take the following quiz. If not, reread from the top.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Pop-gene: Genetic Drift”]
[h]Genetic Drift
[q topic= “genetic_drift”]Change in allele frequencies caused by random selection and reproduction of alleles is called
[c]IG11dGF0aW9uYWwgdmFyaWF0aW9u[Qq]
[c]IG5hdHVyYWwgc2VsZWN0aW9u[Qq]
[c]IHNleHVhbCBzZWxlY3Rpb24=[Qq]
[c]IGdlbmV0aW MgZHJpZnQ=[Qq]
[f]IE5vLiBXaGF04oCZcyBkZXNjcmliZWQgYWJvdmUgaXMgYSByYW5kb20gcHJvY2Vzcywgd2l0aG91dCBkaXJlY3Rpb24uIFRoaW5rIG9mIHdoYXQgYSByYWZ0IGRvZXMgaW4gYSBzd2ltbWluZyBwb29sLCBhbmQgeW914oCZbGwgaGF2ZSB0aGUgYW5zd2VyLg==[Qq]
[f]IE5vLiBOYXR1cmFsIHNlbGVjdGlvbiBpcyBkaXJlY3Rpb25hbC4gV2hhdOKAmXMgZGVzY3JpYmVkIGFib3ZlIGlzIGEgcmFuZG9tIHByb2Nlc3MsIHdpdGhvdXQgZGlyZWN0aW9uLiBUaGluayBvZiB3aGF0IGEgcmFmdCBkb2VzIGluIGEgc3dpbW1pbmcgcG9vbCwgYW5kIHlvdeKAmWxsIGhhdmUgdGhlIGFuc3dlci4=[Qq]
[f]IE5vLiBTZXh1YWwgc2VsZWN0aW9uIGlzIGRpcmVjdGlvbmFsLiBXaGF04oCZcyBkZXNjcmliZWQgYWJvdmUgaXMgYSByYW5kb20gcHJvY2Vzcywgd2l0aG91dCBkaXJlY3Rpb24uIFRoaW5rIG9mIHdoYXQgYSByYWZ0IGRvZXMgaW4gYSBzd2ltbWluZyBwb29sLCBhbmQgeW914oCZbGwgaGF2ZSB0aGUgYW5zd2VyLg==[Qq]
[f]IFllcy4g4oCYR2VuZXRpYyBkcmlmdOKAmSBpcyBhIA==Y2hhbmdlIGluIGFsbGVsZSBmcmVxdWVuY2llcyBjYXVzZWQgYnkgcmFuZG9tIHNlbGVjdGlvbiBhbmQgcmVwcm9kdWN0aW9uIG9mIGFsbGVsZXMu
Cg==[Qq]
[q topic= “genetic_drift”]Genetic drift is
[c]IGludGVudGlvbmFsLg==[Qq]
[c]IHJhbm RvbS4=[Qq]
[c]IGRpcmVjdGlvbmFsIChsaWtlIG5hdHVyYWwgc2VsZWN0aW9uKS4=[Qq]
[f]IE5vLiBGaXJzdGx5LCB0aGUgd29yZCDigJhpbnRlbnRpb25hbOKAmSBkb2VzbuKAmXQgcmVhbGx5IGFwcGx5IHRvIGFueSB0eXBlIG9mIGV2b2x1dGlvbmFyeSBjaGFuZ2UuIEluIHRoZSBjYXNlIG9mIGdlbmV0aWMgZHJpZnQsIGp1c3QgdGhpbmsgb2YgdGhlIHdvcmQg4oCYZHJpZnQs4oCZIGFuZCBob3cgZHJpZnRpbmcgbW92ZXMgeW91IGFyb3VuZC4=[Qq]
[f]IFllcyBBIGtleSBmZWF0dXJlIG9mIGdlbmV0aWMgZHJpZnQgaXMgaXRzIHJhbmRvbW5lc3MuIEluIGdlbmV0aWMgZHJpZnQsIA==Y2hhbmdlIGluIGFsbGVsZSBmcmVxdWVuY2llcyBpcyBjYXVzZWQgYnkgcmFuZG9tIHNlbGVjdGlvbiBhbmQgcmVwcm9kdWN0aW9uIG9mIGFsbGVsZXM=[Qq]
[f]IE5vLiBBIGtleSBmZWF0dXJlIG9mIGdlbmV0aWMgZHJpZnQgaXMgdGhhdCBpdOKAmXMgTk9UIGRpcmVjdGlvbmFsLiBKdXN0IHRoaW5rIG9mIHRoZSB3b3JkIOKAmGRyaWZ0LOKAmSBhbmQgaG93IGRyaWZ0aW5nIG1vdmVzIHlvdSBhcm91bmQsIGFuZCB5b3XigJlsbCBoYXZlIHRoZSBhbnN3ZXIu
Cg==[Qq]
[q topic= “genetic_drift”]Genetic drift most easily happens in _______________ populations.
[c]IHNt YWxs[Qq]
[c]IGxhcmdl[Qq]
[f]IFllcy4gSXTigJlzIG11Y2ggZWFzaWVyIGZvciBnZW5ldGljIGRyaWZ0IHRvIGFsdGVyIGFsbGVsZSBmcmVxdWVuY2llcyBpbiBzbWFsbCBwb3B1bGF0aW9ucy4gSW4gbGFyZ2UgcG9wdWxhdGlvbnMsIHRoZXJl4oCZcyB0b28gbXVjaCByYW5kb20gdmFyaWF0aW9uIGZvciBkcmlmdCB0byBoYXZlIG11Y2ggb2YgYW4gaW1wYWN0IG9uIGFsbGVsZSBmcmVxdWVuY2llcy4=[Qq]
[f]IE5vLiBJdOKAmXMgbXVjaCBlYXNpZXIgZm9yIGdlbmV0aWMgZHJpZnQgdG8gYWx0ZXIgYWxsZWxlIGZyZXF1ZW5jaWVzIGluIHNtYWxsIHBvcHVsYXRpb25zLiBJbiBsYXJnZSBwb3B1bGF0aW9ucywgdGhlcmXigJlzIHRvbyBtdWNoIHJhbmRvbSB2YXJpYXRpb24gZm9yIGRyaWZ0IHRvIGhhdmUgbXVjaCBvZiBhbiBpbXBhY3Qgb24gYWxsZWxlIGZyZXF1ZW5jaWVzLg==
Cg==[Qq]
[q topic= “genetic_drift”]Populations that have experienced genetic drift will most likely have _________ variation than populations that haven’t experienced genetic drift.
[c]IHRoZSBzYW1lIGFtb3VudCBvZg==[Qq]
[c]IG1vcmU=[Qq]
[c]IGxl c3M=[Qq]
[f]IE5vLiBHZW5ldGljIGRyaWZ0LCB3aXRoIGl0cyByYW5kb20gc2VsZWN0aW9uIG9mIGFsbGVsZXMsIG1vc3QgbGlrZWx5IHJlc3VsdHMgaW4gcG9wdWxhdGlvbnMgd2l0aCBMRVNTIHZhcmlhdGlvbiB0aGFuIHBvcHVsYXRpb25zIHRoYXQgaGF2ZW7igJl0IGV4cGVyaWVuY2VkIGdlbmV0aWMgZHJpZnQu[Qq]
[f]IE5vLiBHZW5ldGljIGRyaWZ0LCB3aXRoIGl0cyByYW5kb20gc2VsZWN0aW9uIG9mIGFsbGVsZXMsIG1vc3QgbGlrZWx5IHJlc3VsdHMgaW4gcG9wdWxhdGlvbnMgd2l0aCBMRVNTIHZhcmlhdGlvbiB0aGFuIHBvcHVsYXRpb25zIHRoYXQgaGF2ZW7igJl0IGV4cGVyaWVuY2VkIGdlbmV0aWMgZHJpZnQu[Qq]
[f]IFllcy4gR2VuZXRpYyBkcmlmdCwgd2l0aCBpdHMgcmFuZG9tIHNlbGVjdGlvbiBvZiBhbGxlbGVzLCBtb3N0IGxpa2VseSByZXN1bHRzIGluIHBvcHVsYXRpb25zIHdpdGggTEVTUyB2YXJpYXRpb24gdGhhbiBwb3B1bGF0aW9ucyB0aGF0IGhhdmVu4oCZdCBleHBlcmllbmNlZCBnZW5ldGljIGRyaWZ0Lg==
Cg==[Qq]
[q topic= “genetic_drift”]Populations that have experienced genetic drift will most likely have _________ fixed alleles in their gene pools than populations that haven’t experienced genetic drift.
[c]IHRoZSBzYW1lIGFtb3VudCBvZg==[Qq]
[c]IG1v cmU=[Qq]
[c]IGxlc3M=[Qq]
[f]IE5vLiBHZW5ldGljIGRyaWZ0LCB3aXRoIGl0cyByYW5kb20gc2VsZWN0aW9uIG9mIGFsbGVsZXMsIG1vc3QgbGlrZWx5IHJlc3VsdHMgaW4gcG9wdWxhdGlvbnMgd2l0aCBMRVNTIHZhcmlhdGlvbiB0aGFuIHBvcHVsYXRpb25zIHRoYXQgaGF2ZW7igJl0IGV4cGVyaWVuY2VkIGdlbmV0aWMgZHJpZnQuIExlc3MgdmFyaWF0aW9uIG1lYW5zIE1PUkUgZml4ZWQgYWxsZWxlcy4=[Qq]
[f]IFllcy4gR2VuZXRpYyBkcmlmdCwgd2l0aCBpdHMgcmFuZG9tIHNlbGVjdGlvbiBvZiBhbGxlbGVzLCBtb3N0IGxpa2VseSByZXN1bHRzIGluIHBvcHVsYXRpb25zIHdpdGggTEVTUyB2YXJpYXRpb24gdGhhbiBwb3B1bGF0aW9ucyB0aGF0IGhhdmVu4oCZdCBleHBlcmllbmNlZCBnZW5ldGljIGRyaWZ0LiBMZXNzIHZhcmlhdGlvbiBtZWFucyBNT1JFIGZpeGVkIGFsbGVsZXMu[Qq]
[f]IE5vLiBHZW5ldGljIGRyaWZ0LCB3aXRoIGl0cyByYW5kb20gc2VsZWN0aW9uIG9mIGFsbGVsZXMsIG1vc3QgbGlrZWx5IHJlc3VsdHMgaW4gcG9wdWxhdGlvbnMgd2l0aCBMRVNTIHZhcmlhdGlvbiB0aGFuIHBvcHVsYXRpb25zIHRoYXQgaGF2ZW7igJl0IGV4cGVyaWVuY2VkIGdlbmV0aWMgZHJpZnQuIExlc3MgdmFyaWF0aW9uIG1lYW5zIE1PUkUgZml4ZWQgYWxsZWxlcy4=
Cg==[Qq]
[q topic= “genetic_drift”]Ellis-van Creveld syndrome is a genetic disease whose symptoms include extra fingers and toes, dwarfism, and heart defects. This disease is in very high frequency among the Amish people who live in Eastern Pennsylvania. The most likely explanation for this is
[c]IG5hdHVyYWwgc2VsZWN0aW9uLg==[Qq]
[c]IHJhbmRvbSBtdXRhdGlvbi4=[Qq]
[c]IGEgcG9wdWxhdGlvbiBib3R0bGVuZWNrLg==[Qq]
[c]IHRoZSBmb3VuZG VyIGVmZmVjdC4=[Qq]
[f]IE5vLiBOYXR1cmFsIHNlbGVjdGlvbiBpbnZvbHZlcyB0aGUgZW52aXJvbm1lbnQgc2VsZWN0aW5nIHBoZW5vdHlwZXMgdGhhdCBhcmUgYmVuZWZpY2lhbCwgYW5kIGFsbGVsZSBmcmVxdWVuY2llcyBzaGlmdGluZyBpbiByZXNwb25zZS4gRWxsaXMtdmFuIENyZXZlbGQgc3luZHJvbWUgaXMgZmFyIGZyb20gYmVuZWZpY2lhbC4=[Qq]
[f]IE5vLiBXaGlsZSBzb21lIGRpc2Vhc2VzIGxpa2UgYWNob25kcm9wbGFzaWEgKGFub3RoZXIgZm9ybSBvZiBkd2FyZmlzbSkgYXJlIG1haW50YWluZWQgYnkgbXV0YXRpb24sIHRoaXMgaXMgbm90IGEgY2F1c2Ugb2YgdGhlIGhpZ2ggaW5jaWRlbmNlIG9mIEVsbGlzLXZhbiBDcmV2ZWxkIHN5bmRyb21lIGFtb25nIHRoZSBBbWlzaC4=[Qq]
[f]IE5vLiBXaGlsZSBFbGxpcy12YW4gQ3JldmVsZCBzeW5kcm9tZSBpcyBhbiBleGFtcGxlIG9mIGdlbmV0aWMgZHJpZnQsIHRoZSBoaXN0b3J5IG9mIHRoZSBBbWlzaCBzaG93cyBubyBwb3B1bGF0aW9uIGJvdHRsZW5lY2su[Qq]
[f]IEV4YWN0bHkuIFRoZSBpbmNpZGVuY2Ugb2YgRWxsaXMtdmFuIENyZXZlbGQgc3luZHJvbWUgaXMgYmVzdCBleHBsYWluZWQgYnkgdGhlIOKAmGZvdW5kZXIgZWZmZWN0LuKAmSBBIHNtYWxsIG51bWJlciBvZiBmb3VuZGVycyBicm91Z2h0IHdpdGggdGhlbSB0aGUgYWxsZWxlIGZvciB0aGlzIHN5bmRyb21lLCBhbmQgdGhpcyBhbGxlbGUgaXMgbm93IGluIGhpZ2ggZnJlcXVlbmN5IGluIHRoZSBBbWlzaCBwb3B1bGF0aW9uLg==
Cg==[Qq]
[q topic= “genetic_drift”]In the diagram below, the different colored shapes represent different alleles in populations, which are represented by circles. What process is shown in the diagram?
[c]IG5hdHVyYWwgc2VsZWN0aW9u[Qq]
[c]IHJhbmRvbSBtdXRhdGlvbg==[Qq]
[c]IGEgcG9wdWxhdGlvbiBib3R0bGVuZWNr[Qq]
[c]IHRoZSBmb3VuZG VyIGVmZmVjdA==[Qq]
[f]IE5vLiBOYXR1cmFsIHNlbGVjdGlvbiBpbnZvbHZlcyB0aGUgZW52aXJvbm1lbnQgc2VsZWN0aW5nIHBoZW5vdHlwZXMgdGhhdCBhcmUgYmVuZWZpY2lhbCwgYW5kIGFsbGVsZSBmcmVxdWVuY2llcyBzaGlmdGluZyBpbiByZXNwb25zZS4gSGVyZSB5b3Ugc2VlIGEgbGFyZ2UgcGFyZW50IHBvcHVsYXRpb24sIGFuZCBzZXZlcmFsIG5ldyBwb3B1bGF0aW9ucyBiZWluZyBmb3VuZGVkLiBUd28gb2YgdGhlIG5ldyBwb3B1bGF0aW9ucyBoYXZlIGxlc3MgZGl2ZXJzaXR5IG9mIGFsbGVsZXMgdGhhbiB0aGUgcGFyZW50IHBvcHVsYXRpb24u[Qq]
[f]IE5vLiBJZiB0aGlzIHdlcmUgYSByYW5kb20gbXV0YXRpb24sIHlvdeKAmWQgc2VlIG5ldyBhbGxlbGVzIGFwcGVhcmluZy4gSGVyZSB5b3Ugc2VlIGEgbGFyZ2UgcGFyZW50IHBvcHVsYXRpb24sIGFuZCBzZXZlcmFsIG5ldyBwb3B1bGF0aW9ucyBiZWluZyBmb3VuZGVkLiBUd28gb2YgdGhlIG5ldyBwb3B1bGF0aW9ucyBoYXZlIGxlc3MgZGl2ZXJzaXR5IG9mIGFsbGVsZXMgdGhhbiB0aGUgcGFyZW50IHBvcHVsYXRpb24u[Qq]
[f]IE5vLiBBIHBvcHVsYXRpb24gYm90dGxlbmVjayB3b3VsZCBiZXN0IGJlIGRlcGljdGVkIGJ5IG1hbnkgbWVtYmVycyBvZiB0aGUgcGFyZW50IHBvcHVsYXRpb24gYmVpbmcgd2lwZWQgb3V0LCB3aXRoIG9ubHkgYSBmZXcgc3Vydml2b3JzIGxlZnQuIEhlcmUgeW91IHNlZSBhIGxhcmdlIHBhcmVudCBwb3B1bGF0aW9uLCBhbmQgc2V2ZXJhbCBuZXcgcG9wdWxhdGlvbnMgYmVpbmcgZm91bmRlZC4gVHdvIG9mIHRoZSBuZXcgcG9wdWxhdGlvbnMgaGF2ZSBsZXNzIGRpdmVyc2l0eSBvZiBhbGxlbGVzIHRoYW4gdGhlIHBhcmVudCBwb3B1bGF0aW9uLg==[Qq]
[f]IEV4YWN0bHkuIFdoYXTigJlzIHNob3duIGluIHRoaXMgZGlhZ3JhbSBpcyB0aGUgZm91bmRlciBlZmZlY3QuIFlvdSBzZWUgYSBsYXJnZSBwYXJlbnQgcG9wdWxhdGlvbiwgYW5kIHNldmVyYWwgbmV3IHBvcHVsYXRpb25zIGJlaW5nIGZvdW5kZWQuIFR3byBvZiB0aGUgbmV3IHBvcHVsYXRpb25zIGhhdmUgbGVzcyBkaXZlcnNpdHkgb2YgYWxsZWxlcyB0aGFuIHRoZSBwYXJlbnQgcG9wdWxhdGlvbi4=
Cg==[Qq]
[q topic= “genetic_drift”]What process is shown in the diagram?
[c]IG5hdHVyYWwgc2VsZWN0aW9u[Qq]
[c]IHJhbmRvbSBtdXRhdGlvbg==[Qq]
[c]IGEgcG9wdWxhdGlv biBib3R0bGVuZWNr[Qq]
[c]IHRoZSBmb3VuZGVyIGVmZmVjdA==[Qq]
[f]IE5vLiBOYXR1cmFsIHNlbGVjdGlvbiBpbnZvbHZlcyB0aGUgZW52aXJvbm1lbnQgc2VsZWN0aW5nIHBoZW5vdHlwZXMgdGhhdCBhcmUgYmVuZWZpY2lhbCwgYW5kIGFsbGVsZSBmcmVxdWVuY2llcyBzaGlmdGluZyBpbiByZXNwb25zZS4gSGVyZSB5b3Ugc2VlIGEgbGFyZ2UgcG9wdWxhdGlvbiB0aGF0IGdldHMgcmVkdWNlZCBhbmQgdGhlbiByZWNvdmVycy4gV2hhdCBldmVudCBpbiBwb3B1bGF0aW9uIGdlbmV0aWNzIGhhcyB0aGlzIHBhdHRlcm4/[Qq]
[f]IE5vLiBSYW5kb20gbXV0YXRpb24gYnJpbmdzIGFib3V0IG5ldyBhbGxlbGVzIGluIGEgcG9wdWxhdGlvbi4gVGhlcmXigJlzIG5vIGV2aWRlbmNlIG9mIHRoYXQgaW4gdGhpcyBkaWFncmFtLiBIZXJlIHlvdSBzZWUgYSBsYXJnZSBwb3B1bGF0aW9uIHRoYXQgZ2V0cyByZWR1Y2VkIGFuZCB0aGVuIHJlY292ZXJzLiBXaGF0IGV2ZW50IGluIHBvcHVsYXRpb24gZ2VuZXRpY3MgaGFzIHRoaXMgcGF0dGVybj8=[Qq]
[f]IFllcy4gQSBwb3B1bGF0aW9uIGJvdHRsZW5lY2sgd291bGQ=wqA=YmUgZGVwaWN0ZWQgYnkgbWFueSBtZW1iZXJzIG9mIHRoZSBwYXJlbnQgcG9wdWxhdGlvbiBiZWluZyB3aXBlZCBvdXQsIHdpdGggb25seSBhIGZldyBzdXJ2aXZvcnMgbGVmdC4gVGhlIHN1cnZpdm9ycyB0aGVuIHJlYnVpbGQgdGhlIHBvcHVsYXRpb24uIFRoYXTigJlzIGV4YWN0bHkgd2hhdCB5b3Ugc2VlIGluIHRoaXMgZGlhZ3JhbS4=[Qq]
[f]IE5vLiBUaGUgZm91bmRlciBlZmZlY3Qgd291bGQgYmUgc2hvd24gYnkgYSBmZXcgaW5kaXZpZHVhbHMgYnJhbmNoaW5nIG9mZiBmcm9tIGEgcGFyZW50IHBvcHVsYXRpb24gdG8gZm9ybSBhIG5ldyBwb3B1bGF0aW9uLiBIZXJlIHlvdSBzZWUgYSBsYXJnZSBwb3B1bGF0aW9uIHRoYXQgZ2V0cyByZWR1Y2VkIGFuZCB0aGVuIHJlY292ZXJzLiBXaGF0IGV2ZW50IGluIHBvcHVsYXRpb24gZ2VuZXRpY3MgaGFzIHRoaXMgcGF0dGVybj8=
Cg==[Qq]
[q topic= “genetic_drift”]Genetically, the cheetah is an example of a species that experienced
[c]IHRoZSBmb3VuZGVyIGVmZmVjdC4=[Qq]
[c]IGEgcG9wdWxhdGlvbi Bib3R0bGVuZWNrLg==[Qq]
[f]IE5vLiBXaGlsZSB0aGUgY2hlZXRhaCBnZW5lIHBvb2wgaXMgYW4gZXhhbXBsZSBvZiBnZW5ldGljIGRyaWZ0LCB0aGUgc3BlY2lmaWMgbWVjaGFuaXNtIGRvZXMgbm90IGxvb2sgbGlrZSB0aGUgZm91bmRlciBlZmZlY3Qu[Qq]
[f]IFllcy4gVGhlIGNoZWV0YWjigJlzIGV4dHJlbWUgbGFjayBvZiBnZW5ldGljIGRpdmVyc2l0eSBpbmRpY2F0ZXMgdGhhdCB0aGUgY2hlZXRhaCBleHBlcmllbmNlZCBhIHBvcHVsYXRpb24gYm90dGxlbmVjayBpbiB0aGUgcGFzdCwgd2lwaW5nIG91dCBtb3N0IG9mIHRoZSBnZW5ldGljIHZhcmlhdGlvbiBpbiB0aGUgY2hlZXRhaCBnZW5lIHBvb2wu
Cg==[Qq]
[q topic= “genetic_drift”]One consequence of the population bottleneck experienced by the Northern elephant seal is
[c]IGxvdyBnZW5ldGlj IHZhcmlhYmlsaXR5[Qq]
[c]IGhpZ2ggcmF0ZXMgb2YgbXV0YXRpb24=[Qq]
[c]IGhpZ2ggZ2VuZXRpYyB2YXJpYWJpbGl0eQ==[Qq]
[f]IFllcy4gVGhlIE5vcnRoZXJuIGVsZXBoYW50IHNlYWwsIHRob3VnaCBpdCBoYXMgYmVlbiBzdWNjZXNzZnVsIGluIHJlYnVpbGRpbmcgaXRzIHBvcHVsYXRpb24sIGhhcyBsb3cgZ2VuZXRpYyB2YXJpYWJpbGl0eS4=[Qq]
[f]IE5vLiBUaGVyZeKAmXMgbm8gZXZpZGVuY2UgdGhhdCBtdXRhdGlvbiByYXRlcyBhcmUgYW55IGhpZ2hlciBpbiBOb3J0aGVybiBlbGVwaGFudCBzZWFscyB0aGFuIHRoZXkgYXJlIGluIGFueSBvdGhlciBwb3B1bGF0aW9uLg==[Qq]
[f]IE5vLiBQb3B1bGF0aW9uIGJvdHRsZW5lY2tzLCBzdWNoIGFzIHRoYXQgZXhwZXJpZW5jZWQgYnkgdGhlIE5vcnRoZXJuIEVsZXBoYW50IFNlYWwsIGRvIG5vdCBpbmNyZWFzZSBnZW5ldGljIHZhcmlhYmlsaXR5Lg==[Qq]
[x]
[restart]
[/qwiz]
Links
This is the last tutorial in this series about population genetics. Depending on your teacher’s instructions, choose one of the options below:
- If you’re in an AP Bio or College Bio class, test your mastery of this topic by taking the population genetics cumulative quiz.
- If you’re in a high school class, use the high school biology course menu above to choose another topic.
