1. Introduction
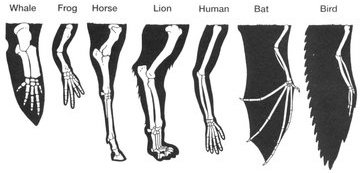
In a previous tutorial in this module, we looked at how homologous structures and vestigial traits serve as evidence for evolution.
|
|
|
In this tutorial, we’ll continue to look at homologies and vestigial traits, but in different places: in embryos, molecules, and genes.
2. Embryology as Evidence for Evolution
The images below show animal embryos at equivalent phases of development. The top row consists of photographs with some modifications (a yolk sac removed in the salamander). The bottom row shows drawings from Ernst Haeckel, a famous 19th century biologist.
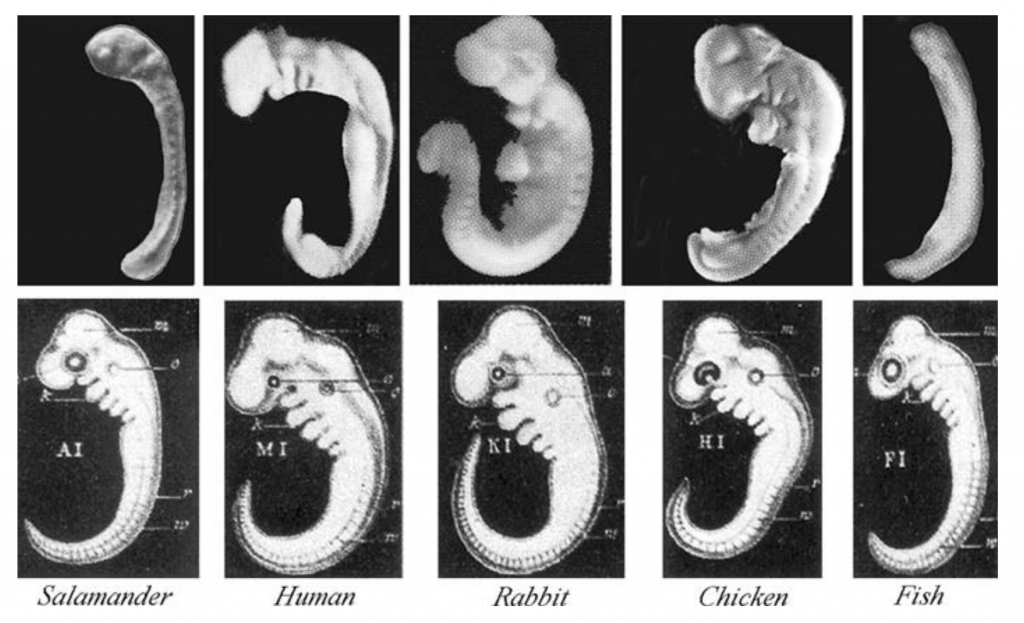
Haeckel created the drawings because he felt that they served as evidence for evolution. Here’s why.
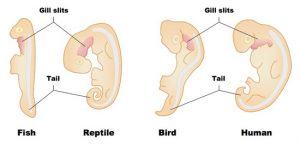
Start by focusing on the column showing a human embryo. There, you can see vestigial traits that are lost as the embryo develops into a fetus. Human embryos have a tail early in development. They also have a series of slits that extend from the neck area below the head into the back of the mouth cavity. In a fish, these slits are maintained in the adult form. They let water flow over the gills, creating a current that enhances absorption of oxygen.
In all tailless vertebrates, like humans and birds, the tail gets broken down through a process called apoptosis: programmed cell death. The material making up the tail gets recycled, and is used to build other parts of the body. The same thing happens to the gill slits in all of the tetrapod (four-limbed) vertebrates. That’s because all of the tetrapods (amphibians included, once they’re adults) absorb oxygen from the air using their lungs, rather than using gills to absorb oxygen from the water.
So why do animals such as humans, rabbits, and chickens, during development, expend the energy to create tails and gill slits, only to break them down and absorb them later? It’s because having a tail (for example) is a part of a developmental plan inherited from a vertebrate ancestor. In early embryonic forms, the genes for this plan are still expressed. The only reason our embryonic form possesses a tail is because it’s part of a developmental program that we inherited from our ancestors. That makes it (along with gill slits) an additional piece of evidence for descent with modification.
Why haven’t the genes for these structures been deleted? We’ll focus on the tail. One possibility is that the developmental plan to build a 4-limbed vertebrate might require that at some moment in development, a tail be present. Without having an embryonic tail, the complex signaling that goes on between cells which results in the various parts of the body growing in the right place could be disrupted. So all vertebrates start life by building a tail, whether they keep it or not. The same is true of the gill slits.
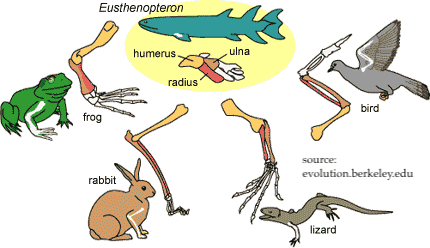
Embryological development, in this sense, is linked to adaptive radiation and homologous structures. All vertebrates descend from a common ancestor. As they develop, their body plan unfolds in a general way common to all vertebrates. As development continues, the pattern diverges in various directions based on the ecological niche and life cycle of the specific lineage. Earliest embryological development expresses the basic vertebrate theme. Then this theme gets varied: A turtle will develop a shell. A bird will develop wings. A human will develop a hand with an opposable thumb.
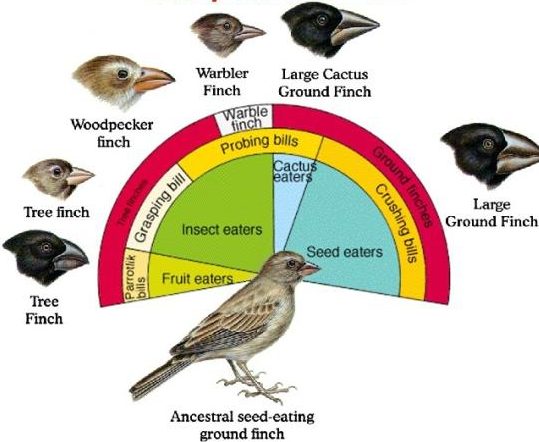 To connect this with a well-known example of adaptive radiation, think of the Galapagos finches. The beaks of early embryonic Galapagos finches are probably very hard to distinguish from one another. But as development unfolds, some beaks grow stouter, for a seed-eating niche, while others grow longer, for probing into flowers. It’s all about homology and descent with modification from a common ancestor.
To connect this with a well-known example of adaptive radiation, think of the Galapagos finches. The beaks of early embryonic Galapagos finches are probably very hard to distinguish from one another. But as development unfolds, some beaks grow stouter, for a seed-eating niche, while others grow longer, for probing into flowers. It’s all about homology and descent with modification from a common ancestor.
3. Molecular Homologies
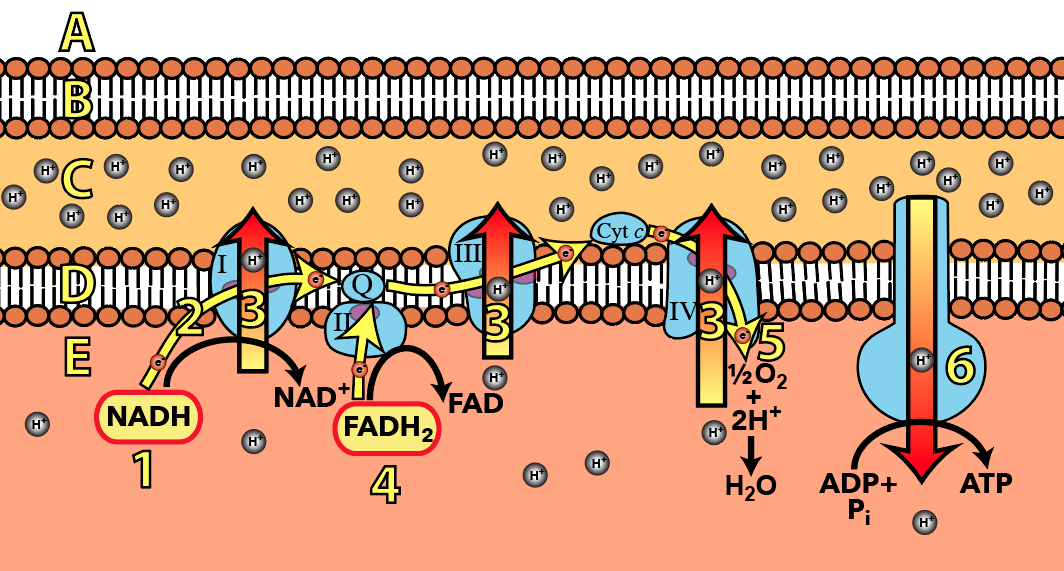
Cytochrome c is a molecule that’s part of the electron transport chain in the inner mitochondrial membrane. It also plays a role in initiating programmed cell death (apoptosis: described above), a process that’s essential to the development of multicellular organisms.
Cytochrome c is found in many species with either an identical or highly similar sequence of amino acids, and a correspondingly similar three dimensional shape. Molecules like cytochrome c — and there are many — are important piece of evidence for evolution. Why do so many different species share this molecule? Because they inherited it from a common ancestor.
To learn more, complete the following interactive reading.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Evidence for evo: Molecular homologies”]
[h]Interactive Reading: Molecular Homologies and Evolution
[i]
[q labels = “top”]Cytochrome c is a protein whose primary structure consists of 104 _________ acids. We’ve previously discussed how the forelimbs of all vertebrates are ______________. The same is true of cytochrome c. Both the forelimb structure and the cytochrome c structures were derived from a common ______________, and _____________ in the descendants.
[l]ancestor
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]amino
[fx] No. Please try again.
[f*] Good!
[l]modified
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l] homologous
[fx] No. Please try again.
[f*] Correct!
[q labels = “top”]The next slide will show a table that shows the number of amino acid differences found between the cytochrome c molecules of human beings and seven other species. In the table, the species names aren’t filled in yet (that’s your job). The more closely __________ a species is to human beings, the fewer _____________ we’d expect to find in the amino acids that make up this protein. Use that hint to complete the table on the next slide.
[l]differences
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l]related
[fx] No. Please try again.
[f*] Good!
[q labels = “right”]Use logic and trial-and-error to complete the table below. Don’t worry about getting a wrong answer.
| Organism | Number of amino acid differences from humans |
| ___________________ | 0 |
| ___________________ | 1 |
| ___________________ | 9 |
| ___________________ | 12 |
| ___________________ | 18 |
| ___________________ | 25 |
| ___________________ | 40 |
[l]Bullfrog
[fx] No. Please try again.
[f*] Good!
[l]chimpanzee
[fx] No. Please try again.
[f*] Correct!
[l]Fruit fly
[fx] No. Please try again.
[f*] Good!
[l]Rabbit
[fx] No. Please try again.
[f*] Great!
[l]Rhesus monkey
[fx] No. Please try again.
[f*] Great!
[l]Pigeon
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l]Yeast
[fx] No, that’s not correct. Please try again.
[f*] Great!
[q]Let’s think about this cytochrome c data. It acts as evidence for evolution because of how it confirms data from other aspects of biology that predated our knowledge of molecular biology. For example, of the other species in this list, chimpanzees are, in terms of their anatomy and behavior, the most [hangman] to humans. Therefore it makes sense that their amino acid sequence for this key protein would be [hangman] to the human sequence. Of the mammals listed, the one that’s the most anatomically and behaviorally different from humans is the [hangman]. It makes sense that this species’ cytochrome c amino acid sequence would, from among the mammals in the table, also be the most [hangman] from the human sequence.
[c]c2ltaWxhcg==[Qq]
[c]aWRlbnRpY2Fs[Qq]
[c]cmFiYml0[Qq]
[c]ZGlmZmVyZW50[Qq]
[q]Why is the pigeon’s cytochrome c sequence more similar to that of humans than is the bullfrog’s? This confirms a similarity based on life cycle. Bullfrogs are amphibians. Their eggs are relatively simple (like fish eggs) and have to develop in the water. The egg of a bird (or a reptile or a mammal) is a much more complex structure. It’s called an amniotic egg, and it contains an internal watery environment (an amniotic sac). This is an adaptation for laying eggs on land. In mammals, the egg has the same structure, but it’s kept inside the body until the embryo has developed into a fetus and then a baby that’s ready to be born.
Based on this adaptation (and others), birds, reptiles, and mammals are all classified as amniotes. So, it makes sense that our cytochrome c would be more similar to that of birds (which are, like us, amniotes), than bullfrogs (which are not).
[q]Another widely conserved molecule among vertebrates is hemoglobin. This is a protein that transports oxygen in red blood cells. The protein consists of four polypeptide chains: two identical alpha chains and two identical beta chains. The alpha chains are 141 amino acids long. The beta chains are 146 amino acids long.
[q labels = “top”]Here’s a list of amino acid differences between the hemoglobin beta chain in humans and the hemoglobin beta chains of several other species.
[l]27
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l]one
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]45
[fx] No. Please try again.
[f*] Good!
[l]eight
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]62
[fx] No. Please try again.
[f*] Excellent!
[q labels = “top”]Let’s see how this confirms our expectations from anatomy, life cycle, etc. Gorillas, like humans, are __________. So it make sense that our hemoglobin is almost __________. Rhesus monkeys are, like us, __________, so it makes sense that their hemoglobin is so similar to ours. Based on hemoglobin, our next closest relative is the ___________, which makes sense because it’s also a _________. The chicken is next: like us it’s a(n) __________. And last is the frog, which is also, like us, a __________.
[l]amniote
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l]apes
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]identical
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]mammal
[fx] No. Please try again.
[f*] Correct!
[l]mouse
[fx] No. Please try again.
[f*] Great!
[l]primates
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]tetrapod
[fx] No, that’s not correct. Please try again.
[f*] Good!
[x]READ THIS CAREFULLY: The conserved nature of proteins like cytochrome c and hemoglobin isn’t unusual. Most proteins that are shared among animals have forms that are in some cases identical, in other cases somewhat modified, but in all cases homologous. That’s why, for example, human diabetics can be treated with insulin that’s derived from animal sources. The insulin produced in the pancreas of a pig only differs from human insulin by one amino acid (out of a total of 21 amino acids in the insulin “A” chain and 30 amino acids in the “B” chain.” That’s true of thousands of proteins.
It’s also true of the genes that code for these proteins, as well as noncoding genes.
Read more below.
[restart]
[/qwiz]
4. Genes can also be homologous and vestigial
Based on how the term “gene” is defined, humans have between 20,000 and 25,000 genes. Almost all of these genes are also found in other animals. In fact, the number of genes that are uniquely human —not found in other animals —might be as low as a few dozen (source: Ed Yong, The Atlantic; also Elizabeth Pennisi, Science).
The thousands of genes that humans share with our closest cousins — the primates, and even other mammals — are clear homologies, Astoundingly, we also share genes with animals with whom our common ancestry stretches back to the origins of animal evolution, over half a billion years ago.
4a. The Hox genes are an ancient set of conserved genes that control development
Like the proteins we discussed above, genes that are shared in a variety of species are called conserved genes. One example of conserved genes shared by almost all animals is a set of genes called the hox genes. These are master control genes that specify the identity of each body segment by controlling what body structure grows on which segment. For example, in insects, a hox gene determines that antennae grow on the head, and that the legs grow on the thorax.
To see how these hox genes have been conserved, start by looking at the image below, and find the line that says “Drosophila HOM-C.”
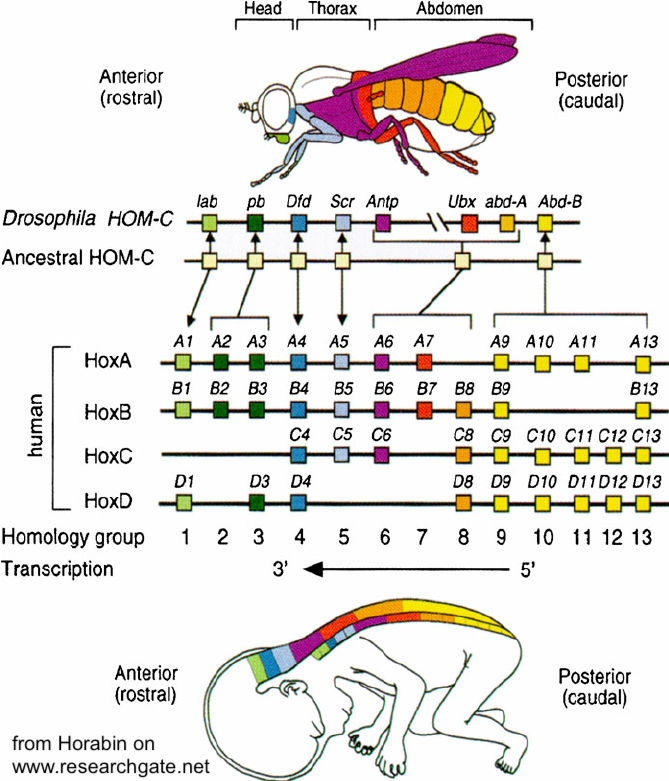
Drosophila melanogaster is a fruit fly. Flies are insects, and insects are arthropods, the group that includes crabs and spiders. These animals have been evolving separately from our group, the vertebrates, for about 600 million years.
In Drosophila, the hox genes are arrayed along the third of Drosophila’s four chromosomes. Each box (“lab,” “pb” etc) on the line labeled “Drosophila Hom-C” represents one of the eight hox genes. Note the matching color between the genes and the body parts. Each hox gene control body parts on one segment. Note how the genes are organized in the sequence as these body parts. The head genes come first, then the front of the thorax, then the back of the thorax, then the abdomen.
Below the “Drosophila” is a line that says “Ancestral.” These are the genes that are thought to have existed in the common ancestor of both arthropods and vertebrates. And below the ancestral genes, you can see how these genes have been transmitted to humans. There’s been duplication of these genes, so that they’re distributed on four chromosomes, but the sequence and function has been conserved. They’re the same genes. Not only are they homologous in terms of DNA nucleotide sequence, but they are transplantable from vertebrate to arthropod. Using the techniques of genetic engineering, a mouse hox gene can be transplanted into a fruit fly. Once there, the mouse hox gene will function to correctly produce the appropriate structure in the right place (for example, an eye on the head).
What’s the takeaway? Conserved genes, like the hox genes, are evidence of descent with modification. Why do humans humans share genes with fruit flies? Because way back in time, we have a common ancestor.
4b. Genes can also be vestigial
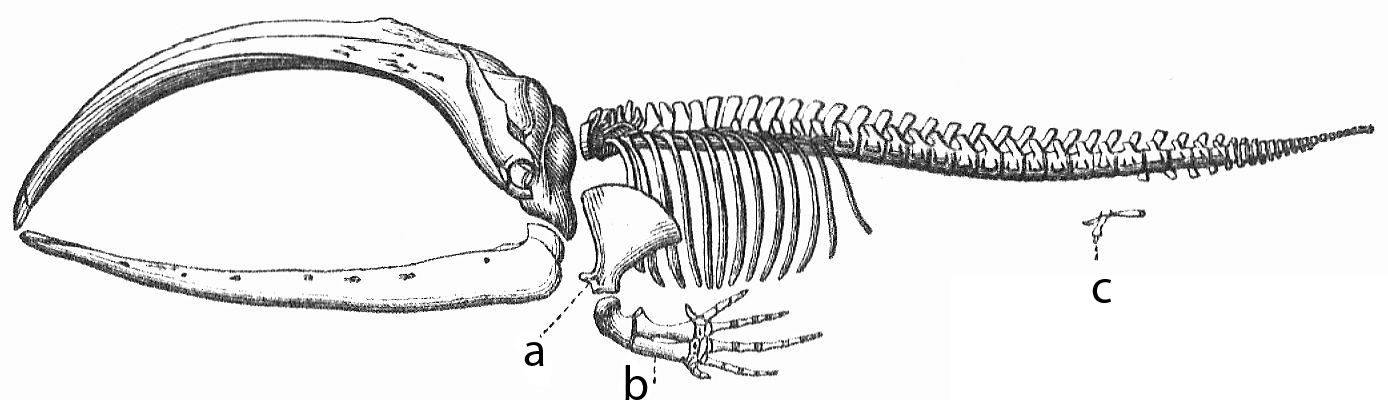
If we think about vestigial structures like the hind limbs of whales, it’s clear that evolution doesn’t necessarily delete the genes for traits that are no longer needed. Rather, the evolutionary process involves diminishing the size of structures (such as the hind limbs of whales), or allowing mutations to accumulate in these structures (such as the vestigial eyes of blind cave fish) so that they’re no longer functional. As Jerry Coyne argues in his book Why Evolution is True, this allows us to make a prediction. In the genomes of many species we should be able to find “silenced or dead genes: genes that once were useful but are no longer intact or expressed.”
The non-functional remnant of a formerly functional gene is called a pseudogene. An example of a pseudogene is the ψGLO (the symbol ψ designates a gene as a pseudogene). The functional GLO gene, in other species, codes for L-Gulonolactone Oxidase. That’s an enzyme that’s used to synthesize Vitamin C (ascorbic acid: C6H8O6)) from glucose (C6H12O6). This enzyme is found in a functional form in almost all mammals, except for bats, guinea pigs, and primates (the order of mammals that includes monkeys, apes, and humans). The groups that don’t make vitamin C have lost the ability to do so because they lived in environments where they could obtain vitamin C from fruit in their environment. Consequently, any mutation in the genes for producing L-Gulonolactone Oxidase would have no negative effect on fitness.
What happens without vitamin C? It took an unintended experiment — sailors undertaking long sea voyages without any available fruit — to see that vitamin C deficiency causes scurvy, the symptoms of which include anemia, weakness, spontaneous bleeding (especially in the gums), loss of teeth, pain in the limbs, and swelling. Prolonged vitamin C deficiency can be fatal. In 1757, a Scottish physician named James Lind persuaded the British Navy to start issuing lime juice to sailors. That ended scurvy in the British Navy, and is why British sailors (and even British people) are sometimes called “limeys.”
About 40 million years ago, an ancient early primate developed a mutation that deactivated the GLO gene. This deactivated gene was passed on to all of his or her descendants, which now includes all primates. With selection no longer maintaining the ψGLO gene’s sequence, the gene could continue to accumulate mutations. In species with extremely recent common ancestors (humans and chimps, for example) a high percentage of mutations are shared. This is called a sequence homology, because it’s a shared sequence from a common ancestor.
In guinea pigs, the mutation that deactivated GLO happened independently. As a result, the sequence of guinea pig ψGLO significantly differs from that in primates (Coyne, 2009)
As with cytochrome c and hemoglobin, ψGLO is not in any way remarkable or unusual. Pseudogenes are common in the human genome. We might have as many pseudogenes as we have genes. In the years since the human genome has been sequenced, the number of pseudogenes that have been identified has come to number in the thousands.
5. Checking Understanding: Embryonic and Molecular Evidence for Evolution
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Evidence for evo: embryonic and molecular homology”]
[h]Embryonic and Molecular Evidence for Evolution
[i]
[q] The embryos of vertebrates such as fish, birds, and mammals develop in similar ways because all of these animals share a common [hangman].
[c]IGFuY2VzdG9y[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] Embryos of humans show various [hangman] traits such as a tail or gill slits that are not present later developmental forms.
[c]IHZlc3RpZ2lhbA==[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] The common developmental pattern shown by various vertebrate animals is evidence of [hangman] with [hangman] from a common ancestor.
[c]IGRlc2NlbnQ=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IG1vZGlmaWNhdGlvbg==[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] Hemoglobin and cytochrome c are both examples of [hangman] at the [hangman] level.
[c]IGhvbW9sb2dpZXM=[Qq]
[f]IENvcnJlY3Qh[Qq]
[c]IG1vbGVjdWxhcg==[Qq]
[f]IEdvb2Qh[Qq]
[q] Based on molecular data like that shown in the table below, one can infer how closely [hangman] two species are.
[c]IHJlbGF0ZWQ=[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] The more similar the amino acid [hangman] in a protein shared by two species, the more closely related they are.
[c]IHNlcXVlbmNl[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q multiple_choice=”true”] Which of the following statements about human genes is most correct?
[c]IEh1bWFucyBhcmUgdW5pcXVlIGJlY2F1c2UgbW9zdCBvZiBvdXIgZ2VuZXMgYXJlIHVuaXF1ZSB3aXRoaW4gdGhlIGFuaW1hbCBraW5nZG9t[Qq]
[f]IE5vLiBPZiB0aGUgMjAsMDAwICYjODIxMTsgMjUsMDAwIGdlbmVzIGluIHRoZSBodW1hbiBnZW5vbWUsIG9ubHkgYSBmZXcgZG96ZW4gYXJlIHVuaXF1ZSB0byBvdXIgc3BlY2llcy4=[Qq]
[c]IEFsbW9zdCBhbGwgaHVtYW4gZ2VuZXMgaGF2ZSBhIGhvbW9s b2dvdXMgY291bnRlcnBhcnQgaW4gb3RoZXIgYW5pbWFscw==[Qq]
[f]IFllcy4gT2YgdGhlIDIwLDAwMCAmIzgyMTE7IDI1LDAwMCBnZW5lcyBpbiB0aGUgaHVtYW4gZ2Vub21lLCBvbmx5IGEgZmV3IGRvemVuIGFyZSB1bmlxdWUgdG8gb3VyIHNwZWNpZXMuIEFsbCB0aGUgb3RoZXJzIGhhdmUgaG9tb2xvZ291cyBjb3VudGVycGFydHMgaW4gb3RoZXIgc3BlY2llcy4=[Qq]
[c]IEFib3V0IGhhbGYgb2YgaHVtYW4gZ2VuZXMgYXJlIHVuaXF1ZSB0byBodW1hbiBiZWluZ3Mu[Qq]
[f]IE5vLsKgT2YgdGhlIDIwLDAwMCAmIzgyMTE7IDI1LDAwMCBnZW5lcyBpbiB0aGUgaHVtYW4gZ2Vub21lLCBvbmx5IGEgZmV3IGRvemVuIGFyZSB1bmlxdWUgdG8gb3VyIHNwZWNpZXMuwqA=[Qq]
[q] Genes or proteins that are widely shared among a variety of species are said to be highly [hangman]. These genes or proteins are molecular [hangman], and indicate common [hangman].
[c]IGNvbnNlcnZlZA==[Qq]
[c]IGhvbW9sb2dpZXM=[Qq]
[c]IGFuY2VzdHJ5[Qq]
[q] The [hangman] genes (shown below) are high level genes involved in development. They control which kind of appendage grows on which body [hangman].
[c]IGhveA==[Qq]
[c]IHNlZ21lbnQ=[Qq]
[q] The similar linear arrangement and sequence similarity in the hox genes indicate that these genes are [hangman]. In addition, the fact that both human and arthropod development is controlled by hox genes indicates that back in time, all animals share a [hangman] [hangman].
[c]IGhvbW9sb2dvdXM=[Qq]
[c]IGNvbW1vbg==[Qq]
[c]IGFuY2VzdG9y[Qq]
[q]The nonfunctional remnant of a formerly functional gene is called a [hangman].
[c]cHNldWRvZ2VuZQ==[Qq]
[q]An example of a pseudogene is ψGLO. Its functional counterpart, the GLO genes, codes for an [hangman] that converts [hangman] into Vitamin C.
[c]ZW56eW1l[Qq]
[c]Z2x1Y29zZQ==[Qq]
[q]In several groups of animals that had easy access to [hangman] in their diet, GLO gene mutated so that it could no longer produce a functional [hangman] for synthesizing vitamin C. However, since vitamin C was available in the environment, the mutation caused no loss of [hangman], allowing the gene to spread throughout the gene pool and eventually become fixed in all populations of primates, guinea pigs, and bats. The nonfunctional remnant of the GLO gene in the genomes of all of these animals is an example of a [hangman].
[c]ZnJ1aXQ=[Qq]
[c]ZW56eW1l[Qq]
[c]Zml0bmVzcw==[Qq]
[c]cHNldWRvZ2VuZQ==[Qq]
[/qwiz]
6. The Deepest Homologies
In biology, the word for a group with a common ancestor is a clade. The Galapagos finches are a clade. So are ants. So are primates.
The shared inheritance of traits from a common ancestor that occurs within a clade is the basis of that clade’s homologous features. In some cases, those homologies are highly conserved (as with molecules like hemoglobin and cytochrome c). In other cases, there’s more flexibility as particular species come to fill specific ecological niches (as with the beaks of the Galapagos finches, or the forelimbs of vertebrates).
Some of the largest groupings of living things are bound together by highly conserved homologies. Let’s think of the largest taxonomic group that we belong to. It’s not the mammals: that’s just our order. It’s not even the animals: that’s our kingdom. It’s our domain, Eukarya.
6a. Commonalities for all eukaryotes

Our domain, the eukaryotes, arose about 1.5 billion years ago. The eukaryotes’ emergence set the stage for life to achieve the complexity associated with multicellular organisms. That includes your complexity and mine, which is embodied in our brains — the most complex object in the universe.
The eukaryotes are a clade. That means that the eukaryotes, despite their incredible diversity, share a single common ancestor. Here’s a short list of some of the conserved homologies that all eukaryotes share.
- Mitochondria
- An endomembrane system: the internal network of membrane-bound organelles such as the rough and smooth endoplasmic reticulum and the Golgi apparatus.
- A membrane-bound nucleus that encloses the cell’s chromosomes, separating the chromosomes from the cytoplasm.
-
Prokaryotic v eukaryotic DNA Linear chromosomes (as opposed to the circular chromosomes in bacteria and archaea).
- Genes that contain introns (intervening segments) that need to be edited out before RNA can be translated into protein.
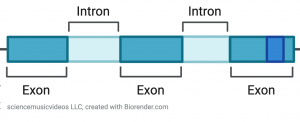
All eukaryotes possess these features. They’re deeply conserved homologies, found in all members of our eukaryotic clade. Note that multicellularity is not one of these features (because there are many eukaryotes that are unicellular).
We can go one level deeper, from traits that unify eukaryotes to conserved traits found in all living things.
6b. Commonalities for all life
All living things possess conserved molecular and genetic homologies that point to one of the profoundest insights from the study of biology: that all living things are related. We’re all cousins. “We,” in this context, doesn’t just mean all human beings (though that’s true). We’re also cousins to dogs, sparrows, salamanders, trout, leaf cutter ants, oyster mushrooms, rhododendrons, kelp, Crenarchaeota (Archaeans that live in hot springs), cyanobacteria (the photosynthetic bacteria that created our oxygen-rich atmosphere) and E. coli (the bacterial that live by the billions in our colons).
To see what these homologies are, and to distinguish the conserved features of all life from the conserved features that bind together the eukaryotes, complete the quiz below.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Evidence for evo: deep homology”]
[h]We’re all cousins
[i]
[q]One conserved homology is seen in the molecule that defines life: the genetic material [hangman]
[c]RE5B[Qq]
[q]All living things follow the central dogma: [hangman] makes [hangman] makes [hangman].
[c]RE5B[Qq]
[c]Uk5B[Qq]
[c]cHJvdGVpbg==[Qq]
[q]All living things synthesize their proteins using [hangman], shown at “2” below.
[c]cmlib3NvbWVz[Qq]
[q]All living things are composed of one or more [hangman].
[c]Y2VsbHM=[Qq]
[q]All living things use [hangman] to power cellular work.
[c]QVRQ[Qq]
[q]This one’s tricky. All living things are made of cells. To be a cell, you have to have a selectively permeable [hangman] surrounding the cytoplasm. But that membrane is not, universally speaking, a [hangman] bilayer (see the diagram at bottom left). That’s because an entire domain of life — Archaea — have a different lipid making up their membrane (see the top molecule below: it has branched side chains, and the glycerol has a different orientation)
[c]bWVtYnJhbmU=[Qq]
[c]cGhvc3Bob2xpcGlk[Qq]
[q]All living things translate their RNA into protein using the same [hangman] [hangman]
[c]Z2VuZXRpYw==[Qq]
[c]Y29kZQ==[Qq]
[q]While all living things have genes that get translated into protein, only the eukaryotes have genes that contain [hangman], intervening sequences that need to be edited out.
[c]aW50cm9ucw==[Qq]
[q]If you had to choose one organelle that defines the eukaryotes, it would probably be the [hangman], because when the bacterial ancestor of these organelles were taken up by an archaeal cell (another type of prokaryote), that’s when the eukaryotes were born.
[c]bWl0b2Nob25kcmlh[Qq]
[q]As opposed to the circular chromosomes in prokaryotes, all eukaryotes have [hangman] chromosomes.
[c]bGluZWFy[Qq]
[q]A conserved homology in all eukaryotes is the presence of an [hangman] system, which includes organelles such as the endoplasmic reticulum and the [hangman] complex.
[c]ZW5kb21lbWJyYW5l[Qq]
[c]Z29sZ2k=[Qq]
[q]In all eukaryotes, the chromosomes are enclosed in a [hangman] that separates the chromosomes from the [hangman].
[c]bnVjbGV1cw==[Qq]
[c]Y3l0b3BsYXNt[Qq]
[x]
[restart]
[/qwiz]
Links
- Click for a cumulative quiz on Evidence for Evolution
- Click to return to the Evidence for Evolution Menu.
Or, use the menus above to choose something else to learn.