1. Watch this video
2a. A few important terms (not in the video) that you should know
- Lineage: a continuous line of descent from a common ancestor to a particular species or group of species. It represents the evolutionary path taken by organisms over time as depicted in a phylogenetic tree.
- Outgroup: a species or group of species that is more distantly related to the organisms being studied (the ingroup). It is used as a point of reference to help determine the evolutionary relationships and shared traits within the ingroup.
- Taxon: (plural: taxa) is any group of organisms that is classified together in a biological classification system. Note that some taxa might be well-formed from an evolutionary perspective, making them clades. But some (however useful) might not make evolutionary sense, such as “waterfowl.”
2b. Study this summary
Phylogeny Basics
- Phylogeny: The evolutionary history of a species or group of species.
- Phylogenetic Tree (Evolutionary Tree): A branching diagram that shows evolutionary relationships, built using morphological (structural), molecular, or genetic evidence.
Key Definitions
Also consult the list of important terms above.
- Clade: A group of organisms that consists of a common ancestor and all its descendants.
- Example: The members of any species are a clade. For example, all humans share a common ancestor.
- Larger clades can include multiple species with shared ancestry. To stick with our lineage, all of the apes — a group including humans, chimps, gorillas, orangutans, and gibbons— are a clade. So are all primates (a larger groups that includes all monkeys, apes, lemurs, and a few other species with traits including large brains, opposable thumbs, fingernails, forward facing eyes, and rotatable shoulder joints).
- Shared Derived Character (or Feature): A trait that evolved in a common ancestor and distinguishes a clade from others.
- Example: Lungs and four limbs are shared derived characters for tetrapods (frogs, lizards, etc.).
- Ancestral Character (or Feature): A trait shared by a clade but also present in larger, more inclusive clades.
- Example: Lungs are ancestral traits for mammals because they are also found in reptiles and amphibians.
Components of Phylogenetic Trees
- Node: A point where two branches diverge, representing a common ancestor.
- Sister Groups: Descendants that diverged from the same node.
- Outgroup: A distantly related group used for comparison to determine evolutionary relationships in the ingroup.
Key Misconceptions
- Vertical closeness (on a horizontally arranged tree, above) does not indicate evolutionary closeness.
- Evolutionary relationships depend on the recency of common ancestry.
- Phylogenetic trees can be rotated around nodes without changing relationships.
Molecular Evidence in Phylogenetics
- Before the 1960s: Morphological similarities were used to create phylogenetic trees.
- Post-1960s: Molecular data (DNA, RNA, and protein sequences) became the gold standard, and resulted in much thinking by biologists about which species were related to one another, and the redrawing of many phylogenies.
- Molecular Clocks: A method to estimate evolutionary timelines based on mutation rates in specific genes or proteins.
- Example: Hemoglobin mutations accumulate at a steady rate, allowing biologists to estimate divergence times among vertebrates.
- Application: Molecular clocks and phylogenetic trees provide insights into evolutionary events, such as the divergence of humans and chimpanzees ~6 million years ago.
3. Master these flashcards
[qdeck bold_text=”false” style=”width: 600px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-Phylogeny, APBVP”]
[h] Phylogeny and Phylogenetic Trees Flashcards
[i]
[start]
[q json=”true” yy=”4″ dataset_id=”Unit 7 Cumulative Flashcards Dataset|4cea5dca74638″ question_number=”37″ unit=”7.Evolution_and_Natural_Selection” topic=”7.9.Phylogeny”] What is phylogeny? What is a phylogenetic tree?
[a] Phylogeny means “evolutionary history.” A phylogenetic tree or evolutionary tree (like the one shown here) is a branching diagram or “tree” showing the evolutionary relationships among various biological species or other taxonomic groups (such as genera or families). Phylogenetic trees are based upon similarities and differences in morphological, molecular, and/or genetic characteristics.
[q] The study of evolutionary history and the relationships among individuals or populations is known as
[a] The study of evolutionary history and the relationships among individuals or populations is known as phylogeny.
[q] In the phylogenetic tree below, letters A, B, C, D, and E represent three things. What are they?
[a] In the diagram to the right
- The letters are branch points (where one species splits into two or more descendants).
- These branch points are also known as nodes.
- Finally, each node is where you find a clade’s common ancestors.
[q] A single line of descent (such as that from “p” to “r”) is known as a _______________.
[a]
A single line of descent is known as a lineage.
[q json=”true” yy=”4″ dataset_id=”Unit 7 Cumulative Flashcards Dataset|4ce8e94305e38″ question_number=”38″ unit=”7.Evolution_and_Natural_Selection” topic=”7.9.Phylogeny”] Define “clade.”
[a] A clade is a group of organisms that consists of a common ancestor and all of that ancestor’s descendants. All of the numbers in the accompanying diagram indicate clades.
[q json=”true” yy=”4″ unit=”7.Evolution_and_Natural_Selection” topic=”7.9.Phylogeny” dataset_id=”Unit 7 Cumulative Flashcards Dataset|4ce72a3a1ae38″ question_number=”39″] Define “shared derived character.”
[a] A shared derived character is a trait that identifies a clade. It evolved in the common ancestor of that clade and sets it apart from other clades. For example, lungs and four limbs are shared derived traits that separate the clade that includes “N” through “S” (four-limbed vertebrates) from group “M” (a fish). The vertebral column is a shared derived character separating the vertebrates (“M” through “S”) from non-vertebrate chordates, such as the hagfish (“L”).
[q json=”true” yy=”4″ unit=”7.Evolution_and_Natural_Selection” topic=”7.9.Phylogeny” dataset_id=”Unit 7 Cumulative Flashcards Dataset|4cdfe394f2638″ question_number=”43″] Define “ancestral feature.”
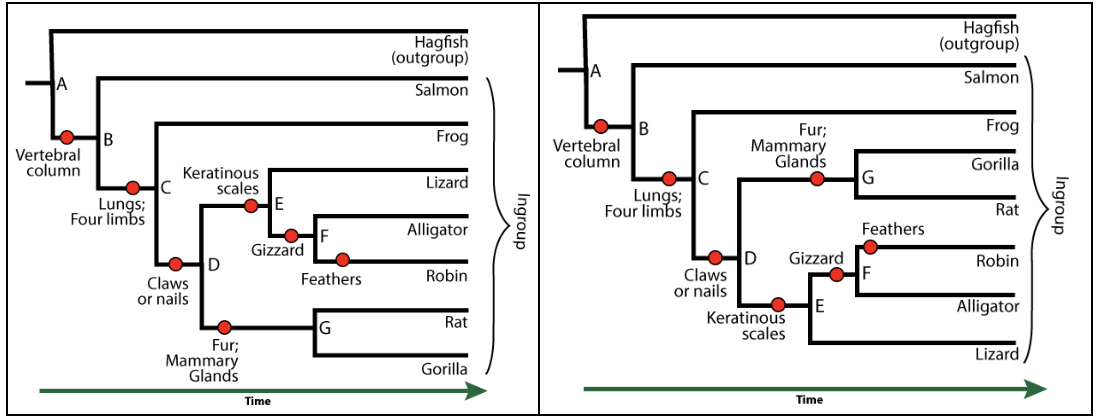
[a] A trait that members of a clade share, but which is also shared by larger, more inclusive clades, is called an ancestral feature. For example, the clade that includes “G” and its descendants (rats and gorillas) is the mammal clade. An ancestral feature of this clade would be “claws or nails.” Mammals have this trait, but it’s also found in other organisms outside the clade (such as birds, alligators, and lizards).
[q] In the phylogenetic tree below, keratinous scales (at “E”) are a(n) __________________ of the clade that includes lizards, alligators, and robins.
[a] Keratinous scales (at “E”) are a shared derived feature of the clade that includes lizards, alligators, and robins.
[q] In the phylogenetic tree below, claws or nails (at “D”) are a(n) __________________ of the clade that includes lizards, alligators, and robins.
[a] Claws or nails (at “D”) are an ancestral feature of the clade that includes lizards, alligators, and robins.
[q] In the phylogenetic tree below, the species that is most closely related to the polar bear is the ___________. Justify your answer.
[a] The species that is most closely related to the polar bear is the brown bear. That’s because the two share the most recent common ancestor.
[q] Why is it incorrect to say that the Giant Panda is most closely related to the Spectacled Bear?
[a] On a phylogenetic tree, closeness is about how recently two taxa share a common ancestor, not who’s next to whom. The giant panda’s most recent common ancestor with the spectacled bear is “A.” That means that the Giant Panda is equally distantly related to every other bear on the tree.
[q] A clade called the ruminantia includes animals such as cows, deer, and antelopes. Their shared derived features include a certain type of hoof and a four-chambered stomach. How would you describe this type of shared derived feature?
[a] These are shared derived features based on anatomy (or body structure). Morphology would also have been a fine answer.
[q] COMPLETE THIS SENTENCE: In a given stretch of DNA or within a protein, the more nucleotides or amino acids that two species share…
[a] In a given stretch of DNA or within a protein, the more nucleotides or amino acids that two species share, the more closely they’re related.
[q] In the DNA sequences shown below, the shared “T” in species E, F, G, and H can be used as a ____________ for classification.
[a] In the DNA sequences shown below, the shared “T” in species E, F, G, and H can be used as a shared derived feature for classification.
[q json=”true” yy=”4″ dataset_id=”Unit 7 Cumulative Flashcards Dataset|4ce5b5b2ac638″ question_number=”40″ unit=”7.Evolution_and_Natural_Selection” topic=”7.9.Phylogeny”] In phylogenetic analysis, what is an outgroup?
[a] When constructing a phylogenetic tree, an outgroup is a more distantly related group of organisms used to determine the evolutionary relationships among the other organisms in the tree (the “ingroup”). The outgroup is a point of comparison for the ingroup. Speaking phylogenetically, the outgroup is a species (or another taxon) that is not a part of the clade to which all of the other organisms in the phylogenetic tree belong. In the accompanying diagram, “A” is the outgroup for clade II.
[q json=”true” yy=”4″ dataset_id=”Unit 7 Cumulative Flashcards Dataset|4ce3f6a9c1638″ question_number=”41″ unit=”7.Evolution_and_Natural_Selection” topic=”7.9.Phylogeny”] On a phylogenetic tree, what are nodes and sister groups?
[a] On a phylogenetic tree, the nodes are where two branches diverge. The nodes, therefore, represent the common ancestor of the two lineages represented by the branches. In the diagram, letters A through E are nodes. Sister groups are the descendants that split apart from the same node (such as the common cactus finch and the large ground finch).
[q json=”true” yy=”4″ dataset_id=”Unit 7 Cumulative Flashcards Dataset|4ce17d5d1f238″ question_number=”42″ unit=”7.Evolution_and_Natural_Selection” topic=”7.9.Phylogeny”] What type of evidence is typically used to construct a phylogenetic tree?
[a] Before the 1960s, classification and phylogenetic analysis was based on morphological similarities between living organisms or fossils. Since the emergence of sequencing technologies in the 1960s and 70s, nucleotide sequences in DNA and RNA (and amino acid sequences in proteins) have become the “gold standard” in determining phylogenetic relationships.
[/qdeck]
4. Tackle these quizzes
4.1. Clades, Phylogeny and Phylogenetic Trees
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Clades, Phylogeny and Phylogenetic Trees, APBVP”]
[h]Clades, Phylogeny and Phylogenetic Trees
[i]
[q] All of the Galapagos finches descend from a common ancestor. That makes the birds below a [hangman]
[c]IGNsYWRl[Qq]
[q] The study of evolutionary history is called [hangman]
[c]IHBoeWxvZ2VueQ==[Qq]
[q] The diagram below says that all bears make up a [hangman].
[c]IGNsYWRl[Qq]
[q] In the diagram below, the shaded rectangles represent _______________. The letters represent common _______________. They’re also called _______________. And because they represent points where one lineage splits into two, they’re also called _______________ points.
[l] ancestors
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l] branch
[fx] No. Please try again.
[f*] Great!
[l] nodes
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l] clades
[fx] No, that’s not correct. Please try again.
[f*] Great!
[q] The common ancestor of clade 6 is
[textentry single_char=”true”]
[c]IE Q=[Qq]
[f]IE5pY2Ugam9iISAmIzgyMjA7RCYjODIyMTsgaXMgdGhlIGNvbW1vbiBhbmNlc3RvciBvZiBjbGFkZSA2Lg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBUaGUgY29tbW9uIGFuY2VzdG9yIG9mIGNsYWRlIDcgaXMgJiM4MjIwO0UuJiM4MjIxOyBJZiB0aGF0JiM4MjE3O3MgdGhlIGNhc2UsIHRoZW4gd2hhdCYjODIxNztzIHRoZSBjb21tb24gYW5jZXN0b3Igb2YgY2xhZGUgNj8=[Qq]
[q] “B” is the common ancestor of which clade?
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IFdheSB0byBnbyEgJiM4MjIwO0ImIzgyMjE7IGlzIHRoZSBjb21tb24gYW5jZXN0b3Igb2YgY2xhZGUgMy4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiAmIzgyMjA7QyYjODIyMTsgaXMgdGhlIGNvbW1vbiBhbmNlc3RvciBvZiBjbGFkZSA1LiBJZiB0aGF0JiM4MjE3O3MgdGhlIGNhc2UsIHRoZW4gJiM4MjIwO0ImIzgyMjE7IGlzIHRoZSBjb21tb24gYW5jZXN0b3Igb2Ygd2hpY2ggY2xhZGU/[Qq]
[q multiple_choice=”true”] How many clades are in this phylogenetic tree?
[c]IDEg[Qq][c]IDUg[Qq][c]IDYg[Qq][c]ID Ex
Cg==[Qq][f]IE5vLiBUaGVyZSYjODIxNztzIG9uZSBjb21tb24gYW5jZXN0b3IsIGJ1dCBtYW55IG1vcmUgY2xhZGVzLg==[Qq]
[f]IE5vLiBUaGVyZSBhcmUgNSBicmFuY2ggcG9pbnRzLCBidXQgbWFueSBtb3JlIGNsYWRlcy4=[Qq]
[f]IE5vLiBUaGVyZSBhcmUgc2l4IGRlc2NlbmRhbnQgc3BlY2llcywgYnV0IG1hbnkgbW9yZSBjbGFkZXMu[Qq]
[f]IE5pY2UhIFRoZXJlIGFyZSAxMSBjbGFkZXMu[Qq]
[q] A diagram like the one shown below is a [hangman] tree.
[c]IHBoeWxvZ2VuZXRpYw==[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] A line of descent from an ancestor to its descendants (which would include “p” through “r” below”) is known as a [hangman].
[c]IGxpbmVhZ2U=[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] In the diagram below, “A” is the [hangman] [hangman] of all of the other species in this phylogenetic tree.
[c]IGNvbW1vbg==[Qq]
[f]IEdyZWF0IQ==[Qq]
[c]IGFuY2VzdG9y[Qq]
[f]IEdyZWF0IQ==[Qq]
[q]Letters “A,” “B,” and “C” can also be thought of as [hangman] points. They’re also called [hangman]. These are points where [hangman] events occurred.
[c]YnJhbmNo[Qq]
[c]bm9kZXM=[Qq]
[c]c3BlY2lhdGlvbg==[Qq]
[/qwiz]
4.2. Phylogenetic Trees
[qwiz style=”width: 700px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-Phylogenetic Trees, APBVP”]
[h] Phylogenetic Trees: Checking Understanding
[i]
[q multiple_choice=”true”] In the phylogenetic tree below, the shared derived trait of clade “F” is (a)
[c]IEtlcmF0aW5vdXMgc2NhbGVz[Qq]
[f]IE5vLiBGb3IgY2xhZGUgJiM4MjIwO0YsJiM4MjIxOyBrZXJhdGlub3VzIHNjYWxlcyB3b3VsZCBiZSBhbiBhbmNlc3RyYWwgZmVhdHVyZS4gQSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIGlzIG9uZSB0aGF0IHNldHMgdGhlIG1lbWJlcnMgb2YgYSBjbGFkZSBhcGFydCBmcm9tIG90aGVyIGNsYWRlcy4=[Qq]
[c]IEdpen phcmQ=[Qq]
[f]IEV4Y2VsbGVudC4gQSBnaXp6YXJkIHNldHMgdGhlIG1lbWJlcnMgb2YgYSBjbGFkZSAmIzgyMjA7RiYjODIyMTsgYXBhcnQgZnJvbSBvdGhlciBjbGFkZXMsIG1ha2luZyBpdCB0aGVpciBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlLg==[Qq]
[c]IEZlYXRoZXJz[Qq]
[f]IE5vLiBPbmx5IHNvbWUgbWVtYmVycyBvZiBjbGFkZSAmIzgyMjA7ZiYjODIyMTsgaGF2ZSBmZWF0aGVycy4gQSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIGlzIG9uZSB0aGF0IGlzIHNoYXJlZCBieSBhbGwgbWVtYmVycyBvZiBhIGNsYWRlLCBhbmQgd2hpY2ggc2V0cyB0aGUgbWVtYmVycyBvZiBhIGNsYWRlIGFwYXJ0IGZyb20gb3RoZXIgY2xhZGVzLiBGaW5kIGEgZmVhdHVyZSB0aGF0IGZpdHMgdGhvc2UgcmVxdWlyZW1lbnRzLg==[Qq]
[q multiple_choice=”true”] In the phylogenetic tree below, an ancestral trait for clade “F” would be
[c]IEtlcmF0aW5v dXMgc2NhbGVz[Qq]
[f]IEV4Y2VsbGVudC4gRm9yIGNsYWRlICYjODIyMDtGLCYjODIyMTsgJiM4MjIwO2tlcmF0aW5vdXMgc2NhbGVzJiM4MjIxOyBpcyBhbiBhbmNlc3RyYWwgZmVhdHVyZS4gSXQgd2FzIGluaGVyaXRlZCBmcm9tIGEgY29tbW9uIGFuY2VzdG9yLCBidXQgbm90IGZyb20gYSBjbG9zZSBlbm91Z2ggYW5jZXN0b3IgdG8gZGlzdGluZ3Vpc2ggdGhpcyBjbGFkZSBmcm9tIG90aGVyIGNsYWRlcy4=[Qq]
[c]IEdpenphcmQ=[Qq]
[f]IE5vLiBBIGdpenphcmQgc2V0cyB0aGUgbWVtYmVycyBvZiBhIGNsYWRlICYjODIyMDtGJiM4MjIxOyBhcGFydCBmcm9tIG90aGVyIGNsYWRlcywgbWFraW5nIGl0IGEgc2hhcmVkIGRlcml2ZWQgZmVhdHVyZS4gQW4gYW5jZXN0cmFsIHRyYWl0IGlzIGFsc28gaW5oZXJpdGVkIGZyb20gYSBjb21tb24gYW5jZXN0b3IsIGJ1dCBpdCYjODIxNztzIG5vdCBhIHRyYWl0IHRoYXQgZGlzdGluZ3Vpc2hlcyB0aGlzIGNsYWRlIGZyb20gb3RoZXIgY2xhZGVzLg==[Qq]
[c]IEZlYXRoZXJz[Qq]
[f]IE5vLiBPbmx5IHNvbWUgbWVtYmVycyBvZiBjbGFkZSAmIzgyMjA7ZiYjODIyMTsgaGF2ZSBmZWF0aGVycywgbWFraW5nIHRoYXQgZmVhdHVyZSBuZWl0aGVyIGEgc2hhcmVkIGRlcml2ZWQgdHJhaXQgbm9yIGFuIGFuY2VzdHJhbCB0cmFpdC4=
Cg==[Qq]
[q] Which of the trees below depicts a taxon that has more than one ancestor?
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IE5pY2Ugam9iISAmIzgyMjA7MyYjODIyMTsgaGFzIG1vcmUgdGhhbiBvbmUgYW5jZXN0b3Iu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJbiB0YXhhIDEgYW5kIDIsIHRoZXJlJiM4MjE3O3MgYSBzaW5nbGUgbm9kZSB0aGF0IGxlYWRzIHRvIGFsbCBvZiB0aGUgZGVzY2VuZGFudHMgd2l0aGluIHRoYXQgY2xhZGUuIFdoaWNoIGRpYWdyYW0gc2hvd3MgbW9yZSB0aGFuIG9uZSBub2RlIGJlbmVhdGggdGhlIGRlc2NlbmRhbnRzPw==[Qq]
[q] Which of the trees below depicts a taxon that’s an incomplete clade?
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IEdvb2Qgam9iISAmIzgyMjA7NCYjODIyMTsgaXMgYW4gaW5jb21wbGV0ZSBjbGFkZSAoYmVjYXVzZSBpdCYjODIxNztzIG1pc3Npbmcgb25lIG9mIGl0cyBkZXNjZW5kYW50cyku[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBBIGxvb2sgZm9yIGEgdGF4b24gaW4gd2hpY2ggdGhlIG1lbWJlcnMgc2hhcmUgdGhlIHNhbWUgY29tbW9uIGFuY2VzdG9yLCBidXQgbm90IGFsbCBvZiB0aGUgZGVzY2VuZGFudHMgYXJlIHBhcnQgb2YgdGhlIGNsYWRlLg==[Qq]
[q] If a taxon consists of an ancestral species and all of its descendants, then it can be described in one word: a [hangman].
[c]IGNsYWRl[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] A key principle of evolutionary classification is the more closely related two taxa are, the more recently they shared a [hangman] [hangman].
[c]IGNvbW1vbg==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IGFuY2VzdG9y[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] In the diagram below, the most recent common ancestor of A and B would be
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IEdyZWF0ISAmIzgyMjA7MSYjODIyMTsgaXMgdGhlIG1vc3QgcmVjZW50IGNvbW1vbiBhbmNlc3RvciBvZiBBIGFuZCBCLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIHRoZSBub2RlIHRoYXQgbW9zdCBpbW1lZGlhdGVseSBjb25uZWN0cyAmIzgyMjA7QSYjODIyMTsgYW5kICYjODIyMDtCLiYjODIyMTsgVGhhdCYjODIxNztzIHRoZWlyIG1vc3QgcmVjZW50IGNvbW1vbiBhbmNlc3Rvci7CoA==[Qq]
[q] In the diagram below, the most recent common ancestor of A, B, C, D, and E would be
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEF3ZXNvbWUhICYjODIyMDsyJiM4MjIxOyBpcyB0aGUgbW9zdCByZWNlbnQgY29tbW9uIGFuY2VzdG9yIG9mIEEsIEIsIEMsIEQsIGFuZCBFLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBNb3ZlIGJhY2sgaW4gdGltZSBmcm9tIEEsIEIsIEMsIEQsIGFuZCBFIGFuZCBmaW5kIGEgcG9pbnQgd2hlcmUgYWxsIHRoZWlyIGxpbmVhZ2VzIGNvbnZlcmdlLiBUaGF0JiM4MjE3O3MgdGhlaXIgY29tbW9uIGFuY2VzdG9yLsKg[Qq]
[q] If you made up a taxon that included A, B, and C, that taxon would not be a [hangman].
[c]IGNsYWRl[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q multiple_choice=”true”] According to the diagram below, which of the following statements is true?
[c]IEh1bWFucyBldm9sdmVkIGZyb20gT2xkIFdvcmxkIG1vbmtleXM=[Qq]
[f]IE5vLiAmIzgyMjA7RXZvbHZlZCBmcm9tJiM4MjIxOyBtZWFucywgbW9yZSBvciBsZXNzLCAmIzgyMjA7YXJlIGRlc2NlbmRlZCBmcm9tLiYjODIyMTsgSHVtYW5zIGFuZCBPbGQgV29ybGQgTW9ua2V5cyBhcmUgZXZvbHV0aW9uYXJ5IA==Y291c2lucw==LiBZb3VyIGNvdXNpbnMgYXJlbiYjODIxNzt0IHlvdXIgYW5jZXN0b3JzLg==[Qq]
[c]IEh1bWFucyBhbmQgbmV3IHdvcmxkIG1vbmtl eXMgYmVsb25nIHRvIHRoZSBzYW1lIGNsYWRl[Qq]
[f]IFRoYXQmIzgyMTc7cyBjb3JyZWN0LiBIdW1hbnMgYW5kIG5ldyB3b3JsZCBtb25rZXlzIHNoYXJlIGEgY29tbW9uIGFuY2VzdG9yIGFuZCBhcmUgdGhlcmVmb3JlIG1lbWJlcnMgb2YgdGhlIHNhbWUgY2xhZGUu[Qq]
[q] Letters “A,” “B,” and “C” are points where [hangman] events occurred. They’re called [hangman].
[c]IHNwZWNpYXRpb24=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IG5vZGVz[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] The basic idea behind molecular clocks is that for any specific protein or nucleic acid, the rate of change caused by accumulated [hangman] is constant over time.
[c]IG11dGF0aW9ucw==[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] Because mutations accumulate in proteins or nucleic acids at a relatively constant rate, you can infer that mice share a more recent common ancestor with [hangman] and gorillas than they do with chickens.
[c]IG1vbmtleXM=[Qq]
[f]IEdvb2Qh[Qq]
[q] In the diagram below, the shared derived trait of clade G is indicated by number
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzQmIzgyMjE7IHNob3dzIHRoZSBzaGFyZWQgZGVyaXZlZCB0cmFpdCBvZiByYXRzIGFuZCBnb3JpbGxhcyAobWVtYmVycyBvZiB0aGUgbWFtbWFsIGNsYWRlKQ==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHRyYWl0IHRoYXQmIzgyMTc7cyB1bmlxdWUgZm9yIHRoZSBjbGFkZSBpbiB3aGljaCBib3RoIHJhdHMgYW5kIGdvcmlsbGFzIGFyZSBmb3VuZC4=[Qq]
[q] In the diagram below, the common ancestor of lizards, alligators, robins, rats, and gorillas is indicated by letter
[textentry single_char=”true”]
[c]IE Q=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwO0QmIzgyMjE7IGlzIHRoZSBjb21tb24gYW5jZXN0b3Igb2YgdGhlIGNsYWRlIHRoYXQgdW5pdGVzIGxpemFyZHMsIGFsbGlnYXRvcnMsIHJvYmlucywgcmF0cywgYW5kIGdvcmlsbGFzLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGb2xsb3cgdGhlIGxpbmVhZ2VzIGxlYWRpbmcgdG8gbGl6YXJkcywgYWxsaWdhdG9ycywgcm9iaW5zLCByYXRzLCBhbmQgZ29yaWxsYXMgYmFjayBpbiB0aW1lIHVudGlsIHRoZXkgbWVldCBpbiBhIGNvbW1vbiBhbmNlc3Rvci4=[Qq]
[q] In the diagram below, what number indicates the most recent ancestral trait of lizards, alligators, robins, rats, and gorillas?
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzImIzgyMjE7IGlzIHRoZSBtb3N0IHJlY2VudCBhbmNlc3RyYWwgdHJhaXQgb2YgdGhlIGNsYWRlIHRoYXQgdW5pdGVzIGxpemFyZHMsIGFsbGlnYXRvcnMsIHJvYmlucywgcmF0cywgYW5kIGdvcmlsbGFzICh0aGUgc2hhcmVkIGRlcml2ZWQgdHJhaXQgaXMgJiM4MjIwOzMuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIHRoZSBzaGFyZWQgZGVyaXZlZCB0cmFpdCBvciB0aGUgY2xhZGUgdGhhdCB1bml0ZXMgbGl6YXJkcywgYWxsaWdhdG9ycywgcm9iaW5zLCByYXRzLCBhbmQgZ29yaWxsYXMuIFRoZW4gZmluZCB0aGUgdHJhaXQgdGhhdCB3b3VsZCBpbmNsdWRlIHRoZSBvdXRncm91cCBvZiB0aGF0IGNsYWRlICh3aGljaCBpcyB0aGUgZnJvZyku[Qq]
[/qwiz]
4.3. Phylogeny Quiz 3
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Phylogeny Quiz 3, APBVP”]
[h]Phylogeny Quiz 3
[i]
[q json=”true” multiple_choice=”true” unit=”7.Evolution” topic=”7.9.Phylogeny” dataset_id=”2019 AP Bio Dataset|1d2bd37e802f30″ question_number=”376″] Which of the following statements about the phylogenetic tree below is correct?
[c]IFRheG9uIDEgKGJpcmRzKSBpcyBub3QgYSBjbGFkZSBiZWNhdXNlIGl0IGV4Y2x1ZGVzIHRheG9uIDIu[Qq]
[f]IE5vLiBBIGNsYWRlIGlzIGRlZmluZWQgYXMgYSBncm91cCB0aGF0IGluY2x1ZGVzIHRoZSBjb21tb24gYW5jZXN0b3IgYW5kIGFsbCBvZiBpdHMgZGVzY2VuZGFudHMuIEJhc2VkIG9uIHRoaXMgcGh5bG9nZW55LCBiaXJkcyBtYWtlIHVwIGEgY2xhZGUu[Qq]
[c]ICYjODIyMDtNYW1tYWxzJiM4MjIxOyBpcyBub3QgYSBjbGFkZSBiZWNhdXNlIHRlbXBvcmFsIGZlbmVzdHJhIGlzIHRoZSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIG9mIG1hbW1hbHMsIGFuZCB0YXhvbiA2IHNoYXJlcyB0aGlzIHRyYWl0Lg==[Qq]
[f]IE5vLiBBIA==c2hhcmVkIGRlcml2ZWQgZmVhdHVyZQ==IGlzIG9uZSB0aGF0JiM4MjE3O3MgZm91bmQgc29sZWx5IHdpdGhpbiB0aGUgbWVtYmVycyBvZiBhIGNsYWRlLiBIYWlyIGFuZCBtYW1tYXJ5IGdsYW5kcyBhcmUgYSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIHRoYXQgdW5pdGVzIG1hbW1hbHMuIFRlbXBvcmFsIGZlbmVzdHJhIGlzIGZvdW5kIGluIG1hbW1hbHMsIGJ1dCBhbHNvIGluIG1vcmUgaW5jbHVzaXZlIGdyb3VwcyAoc3VjaCBhcyB0aGUgY2xhZGUgbWFkZSB1cCBvZiB0YXhvbiA2IGFuZCB0aGUgdHdvIG1hbW1hbCB0YXhhKS4gVGhhdCBtYWtlcyBpdCBhbiA=YW5jZXN0cmFsIGZlYXR1cmU=IG9mIG1hbW1hbHMu[Qq]
[c]ICYjODIyMDtSZXB0aWxlcyYjODIyMTsgaXMgbm90IGEgY2xhZG UgYmVjYXVzZSBpdCBleGNsdWRlcyB0YXhvbiAxIChiaXJkcyku[Qq]
[f]IE5pY2UuIEEgY2xhZGUgaXMgZGVmaW5lZCBhcyBhIGdyb3VwIHRoYXQgaW5jbHVkZXMgdGhlIGNvbW1vbiBhbmNlc3RvciBhbmQgYWxsIG9mIGl0cyBkZXNjZW5kYW50cy4gQmFzZWQgb24gdGhpcyBwaHlsb2dlbnksIHJlcHRpbGVzIGFyZSBub3QgYSBjbGFkZSBiZWNhdXNlIHRoZXkgZXhjbHVkZSBvbmUgb2YgdGhlIHRheGEgd2l0aCB3aGljaCB0aGV5IHNoYXJlIGEgY29tbW9uIGFuY2VzdHJ5LCB0aGUgYmlyZHMu[Qq]
[c]ICYjODIyMDtNYW5kaWJ1bGFyIGZlbmVzdHJhJiM4MjIxOyBpcyBhbiBhbmNlc3RyYWwgZmVhdHVyZSBvZiB0aGUgY2xhZGUgbWFkZSBvZiB0YXhhIDEgYW5kIDIu[Qq]
[f]IE5vLiBBIA==c2hhcmVkIGRlcml2ZWQgZmVhdHVyZQ==IGlzIG9uZSB0aGF0JiM4MjE3O3MgZm91bmQgc29sZWx5IHdpdGhpbiB0aGUgbWVtYmVycyBvZiBhIGNsYWRlLiBUaGF0IG1ha2VzIG1hbmRpYnVsYXIgZmVuZXN0cmEgYSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIG9mIHRoZSBjbGFkZSBtYWRlIG9mIHRheGEgMSBhbmQgMiwgcmF0aGVyIHRoYW4gYW4gYW5jZXN0cmFsIGZlYXR1cmUu
Cg==[Qq]
[q json=”true” multiple_choice=”true” unit=”7.Evolution” topic=”7.9.Phylogeny” dataset_id=”2019 AP Bio Dataset|1d2a9484232b30″ question_number=”377″] Which of the following statements about the phylogenetic tree below is correct?
[c]IE1hbmRpYnVsYXIgZmVuZXN0cmEgaXMgYSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIG9mIHRoZSBjbGFkZSBjb2 1wb3NlZCBvZiB0YXhhIDEgYW5kIDIuIEFuIGFtbmlvdGljIGVnZyBpcyBhbiBhbmNlc3RyYWwgZmVhdHVyZS4=[Qq]
[f]IEV4Y2VsbGVudC4gQSA=c2hhcmVkIGRlcml2ZWQgZmVhdHVyZQ==IGlzIG9uZSB0aGF0JiM4MjE3O3MgZm91bmQgc29sZWx5IHdpdGhpbiB0aGUgbWVtYmVycyBvZiBhIGNsYWRlLiBUaGF0IG1ha2VzIG1hbmRpYnVsYXIgZmVuZXN0cmEgYSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIG9mIHRoZSBjbGFkZSBtYWRlIG9mIHRheGEgMSBhbmQgMi4gQW4gYW5jZXN0cmFsIGZlYXR1cmUgaXMgZm91bmQgd2l0aGluIG1lbWJlcnMgb2YgYSBjbGFkZSBhbmQgYWxzbyBpbiBtb3JlIGluY2x1c2l2ZSBjbGFkZXMuIFRoYXQgbWFrZXMgYW4gYW1uaW90aWMgZWdnIGFuIGFuY2VzdHJhbCBmZWF0dXJlIG9mIHRoZSBjbGFkZSBtYWRlIG9mIHRheGEgMSBhbmQgMi4=[Qq]
[c]IE1hbmRpYnVsYXIgZmVuZXN0cmEgaXMgYSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIG9mIHRoZSBjbGFkZSBjb21wb3NlZCBvZiB0YXhhIDEgYW5kIDIuIFRlbXBvcmFsIGZlbmVzdHJhIGlzIGFuIGFuY2VzdHJhbCBmZWF0dXJlLg==[Qq]
[f]IE5vLiBUaGUgcHJvYmxlbSBpcyB3aXRoIHRoZSBzZWNvbmQgcGFydCBvZiB0aGUgc3RhdGVtZW50LiBUZW1wb3JhbCBmZW5lc3RyYSBpcyBuZWl0aGVyIGEgc2hhcmVkIGRlcml2ZWQgZmVhdHVyZSBub3IgYW4gYW5jZXN0cmFsIGZlYXR1cmUgb2YgdGhlIGNsYWRlIG1hZGUgb2YgdGF4YSAxIGFuZCAyLg==[Qq]
[c]IEFuIGFtbmlvdGljIGVnZyBpcyBhbiBhbmNlc3RyYWwgZmVhdHVyZSBvZiB0aGUgY2xhZGUgY29tcG9zZWQgb2YgdGF4YSAxIHRocm91Z2ggOC4=[Qq]
[f]IE5vLiBBbiBhbmNlc3RyYWwgZmVhdHVyZSBpcyBmb3VuZCB3aXRoaW4gbWVtYmVycyBvZiBhIGNsYWRlIGFuZCBhbHNvIGluIG1vcmUgaW5jbHVzaXZlIGNsYWRlcy4gQSA=c2hhcmVkIGRlcml2ZWQgZmVhdHVyZQ==IGlzIG9uZSB0aGF0JiM4MjE3O3MgZm91bmQgc29sZWx5IHdpdGhpbiB0aGUgbWVtYmVycyBvZiBhIGNsYWRlLiBUaGF0IG1ha2VzIGhhdmluZyBhbiBhbW5pb3RpYyBlZ2cgYSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIG9mIHRoZSBjbGFkZSBjb21wb3NlZCBvZiB0YXhhIDEgdGhyb3VnaCA4Lg==[Qq]
[c]IEFuIGFtbmlvdGljIGVnZyBpcyBhIHNoYXJlZCBkZXJpdmVkIGZlYXR1cmUgb2YgdGhlIGNsYWRlIGNvbXBvc2VkIG9mIHRheGEgMSBhbmQgMi4gTWFuZGlidWxhciBmZW5lc3RyYSBpcyBhbiBhbmNlc3RyYWwgZmVhdHVyZSB0aGF0IGNsYWRlLg==[Qq]
[f]IE5vLiBBIA==c2hhcmVkIGRlcml2ZWQgZmVhdHVyZQ==IGlzIG9uZSB0aGF0JiM4MjE3O3MgZm91bmQgc29sZWx5IHdpdGhpbiB0aGUgbWVtYmVycyBvZiBhIGNsYWRlLiBBbiBhbmNlc3RyYWwgZmVhdHVyZSBpcyBmb3VuZCB3aXRoaW4gbWVtYmVycyBvZiBhIGNsYWRlIGFuZCBhbHNvIGluIG1vcmUgaW5jbHVzaXZlIGNsYWRlcy4=[Qq]
[q json=”true” multiple_choice=”true” unit=”7.Evolution” topic=”7.9.Phylogeny” dataset_id=”2019 AP Bio Dataset|1d28d57b382b30″ question_number=”378″] A team of biologists is evaluating the phylogenetic tree below.
One biologist suggests that because of similarities between species in taxon 2 and taxon 6, the phylogenetic tree should be revised. Which of the following would be the strongest evidence for keeping taxon 2 and taxon 6 in their current position?
[c]IERldGFpbGVkIGFuYWx5c2lzIG9mIHRoZSBib25lIHN0cnVjdHVyZSBvZiBmb3NzaWwgc3BlY2llcyBmcm9tIHRoZSB0d28gdGF4YS4=[Qq]
[f]IE5vLiBXaGVuIGNvbnN0cnVjdGluZyBhIHBoeWxvZ2VuZXRpYyB0cmVlLCB5b3UgbmVlZCBzb21ldGhpbmcgdGhhdCBsZXRzIHlvdSBjb25jbHVzaXZlbHkgZGlzdGluZ3Vpc2ggaG9tb2xvZ3kgKHdoaWNoIGNvbWVzIGZyb20gY29tbW9uIGFuY2VzdHJ5KSBmcm9tIGFuYWxvZ3kgKHdoaWNoIGNvbWVzIGZyb20gY29udmVyZ2VudCBldm9sdXRpb24pLiBXaGlsZSBib25lIHN0cnVjdHVyZSBjYW4gYmUgdXNlZnVsLCB0aGVyZSYjODIxNztzIGEgYmV0dGVyIHNvdXJjZSBvZiBpbmZvcm1hdGlvbi4=[Qq]
[c]IFgtcmF5IGNyeXN0YWxsb2dyYXBoeSBzdHVkaWVzIHRoYXQgc2hvdyBzaW1pbGFyaXRpZXMgYW5kIGRpZmZlcmVuY2VzIGluIGVuenltZXMgd2l0aGluIHNoYXJlZCBtZXRhYm9saWMgcGF0aHdheXMu[Qq]
[f]IE5vLiBXaGVuIGNvbnN0cnVjdGluZyBhIHBoeWxvZ2VuZXRpYyB0cmVlLCB5b3UgbmVlZCBzb21ldGhpbmcgdGhhdCBsZXRzIHlvdSBjb25jbHVzaXZlbHkgZGlzdGluZ3Vpc2ggaG9tb2xvZ3kgKHdoaWNoIGNvbWVzIGZyb20gY29tbW9uIGFuY2VzdHJ5KSBmcm9tIGFuYWxvZ3kgKHdoaWNoIGNvbWVzIGZyb20gY29udmVyZ2VudCBldm9sdXRpb24pLiBXaGlsZSBlbnp5bWUgc3RydWN0dXJlIGNhbiBiZSB1c2VmdWwsIHRoZXJlJiM4MjE3O3MgYSBiZXR0ZXIgc291cmNlIG9mIGluZm9ybWF0aW9uLg==[Qq]
[c]IEFuYWx5c2lzIG9mIHNlcXVlbmNlcyBvZiBhbW lubyBhY2lkcyBpbiBzaGFyZWQgcHJvdGVpbnMu[Qq]
[f]IEdyZWF0IGpvYi4gU2hhcmVkIHNlcXVlbmNlcyBvZiB0aGUgYW1pbm8gYWNpZHMgaW4gYSBwcm90ZWluIG9yIGluIHRoZSBudWNsZW90aWRlcyBpbiBudWNsZWljIGFjaWRzIGFyZSB0aGUgJiM4MjIwO2dvbGQgc3RhbmRhcmQmIzgyMjE7IGZvciBkaXN0aW5ndWlzaGluZyBiZXR3ZWVuIGhvbW9sb2d5ICh3aGljaCBjb21lcyBmcm9tIGNvbW1vbiBhbmNlc3RyeSkgZnJvbSBhbmFsb2d5ICh3aGljaCBjb21lcyBmcm9tIGNvbnZlcmdlbnQgZXZvbHV0aW9uKS4gU2luY2UgcGh5bG9nZW5ldGljIHRyZWVzIGFyZSBhbGwgYWJvdXQgZGVwaWN0aW5nIHBhdHRlcm5zIG9mIGV2b2x1dGlvbiBiYXNlZCBvbiBjb21tb24gYW5jZXN0cnksIG1vbGVjdWxhciBzZXF1ZW5jZSBkYXRhIGlzIGFsd2F5cyBwcmVmZXJyZWQgb3ZlciBtb3JwaG9sb2dpY2FsIGRhdGEu[Qq]
[c]IFJhZGlvYWN0aXZlIGRhdGluZyBvZiBmb3NzaWxzIG9mIHNwZWNpZXMgZnJvbSB0aGUgdHdvIGdyb3VwcyBzaG93aW5nIHdoZW4gdGhleSBkaXZlcmdlZCBmcm9tIGEgY29tbW9uIGFuY2VzdG9yLg==[Qq]
[f]IE5vLiBXaGVuIGNvbnN0cnVjdGluZyBhIHBoeWxvZ2VuZXRpYyB0cmVlLCB5b3UgbmVlZCBzb21ldGhpbmcgdGhhdCBsZXRzIHlvdSBjb25jbHVzaXZlbHkgZGlzdGluZ3Vpc2ggaG9tb2xvZ3kgKHdoaWNoIGNvbWVzIGZyb20gY29tbW9uIGFuY2VzdHJ5KSBmcm9tIGFuYWxvZ3kgKHdoaWNoIGNvbWVzIGZyb20gY29udmVyZ2VudCBldm9sdXRpb24pLiBTZWUgaWYgeW91IGNhbiBpZGVudGlmeSBhIGZlYXR1cmUgdGhhdCBsZXRzIHlvdSBkbyB0aGF0Lg==
Cg==[Qq]
[q json=”true” multiple_choice=”true” unit=”7.Evolution” topic=”7.9.Phylogeny” dataset_id=”2019 AP Bio Dataset|1d2791d8c35f30″ question_number=”379″] How many clades are in the phylogenetic tree below?
[c]IDEg[Qq][c]IDUg[Qq][c]IDYg[Qq][c]ID Ex
Cg==[Qq][f]IE5vLiBBIGNsYWRlIGlzIGEgZ3JvdXAgdGhhdCBjb25zaXN0cyBvZiBhIGNvbW1vbiBhbmNlc3RvciBhbmQgYWxsIG9mIGl0cyBkZXNjZW5kYW50cy4gSW4gdGhlIHRyZWUgYWJvdmUsIHRoZXJlJiM4MjE3O3Mgb25lIGNvbW1vbiBhbmNlc3RvciwgYnV0IG1hbnkgbW9yZSBjbGFkZXMu
Cg==SGVyZSYjODIxNztzIGEgaGludC4gVGhlIGRpYWdyYW0gYmVsb3cgc2hvd3MgdGhlIHNhbWUgcGh5bG9nZW5ldGljIHRyZWUgYXMgdGhlIGRpYWdyYW0gYWJvdmUsIGJ1dCB3aXRoIHNoYWRpbmcuIE5vdGUgdGhhdCBlYWNoIHNwZWNpZXMgaXMgYSBjbGFkZS4gU28gaXMgZWFjaCBzaGFkZWQgcmVjdGFuZ2xlLiBJZiBhIHNoYWRlZCByZWN0YW5nbGUgaXMgYSBjbGFkZSB3aXRoIG9ubHkgb25lIHNwZWNpZXMgKHN1Y2ggYXMgd2l0aCB0aGUgR3JlZW4gV2FyYmxlciBGaW5jaCBvciB0aGUgVmVnZXRhcmlhbiBGaW5jaCksIGNvdW50IGl0IG9ubHkgb25jZS4=
Cg==[Qq]
[f]IE5vLiBBIGNsYWRlIGlzIGEgZ3JvdXAgdGhhdCBjb25zaXN0cyBvZiBhIGNvbW1vbiBhbmNlc3RvciBhbmQgYWxsIG9mIGl0cyBkZXNjZW5kYW50cy4gSW4gdGhlIHRyZWUgYWJvdmUsIHRoZXJlIGFyZSA1IGJyYW5jaCBwb2ludHMsIGJ1dCBtYW55IG1vcmUgY2xhZGVzLg==
Cg==SGVyZSYjODIxNztzIGEgaGludC4gVGhlIGRpYWdyYW0gYmVsb3cgc2hvd3MgdGhlIHNhbWUgcGh5bG9nZW5ldGljIHRyZWUgYXMgdGhlIGRpYWdyYW0gYWJvdmUsIGJ1dCB3aXRoIHNoYWRpbmcuIE5vdGUgdGhhdCBlYWNoIHNwZWNpZXMgaXMgYSBjbGFkZS4gU28gaXMgZWFjaCBzaGFkZWQgcmVjdGFuZ2xlLiBJZiBhIHNoYWRlZCByZWN0YW5nbGUgaXMgYSBjbGFkZSB3aXRoIG9ubHkgb25lIHNwZWNpZXMgKHN1Y2ggYXMgd2l0aCB0aGUgR3JlZW4gV2FyYmxlciBGaW5jaCBvciB0aGUgVmVnZXRhcmlhbiBGaW5jaCksIGNvdW50IGl0IG9ubHkgb25jZS4=
Cg==[Qq]
[f]IE5vLiBBIGNsYWRlIGlzIGEgZ3JvdXAgdGhhdCBjb25zaXN0cyBvZiBhIGNvbW1vbiBhbmNlc3RvciBhbmQgYWxsIG9mIGl0cyBkZXNjZW5kYW50cy4gSW4gdGhlIHRyZWUgYWJvdmUsIHRoZXJlIGFyZSBzaXggZGVzY2VuZGFudCBzcGVjaWVzLCBidXQgbWFueSBtb3JlIGNsYWRlcy4=
Cg==SGVyZSYjODIxNztzIGEgaGludC4gVGhlIGRpYWdyYW0gYmVsb3cgc2hvd3MgdGhlIHNhbWUgcGh5bG9nZW5ldGljIHRyZWUgYXMgdGhlIGRpYWdyYW0gYWJvdmUsIGJ1dCB3aXRoIHNoYWRpbmcuIE5vdGUgdGhhdCBlYWNoIHNwZWNpZXMgaXMgYSBjbGFkZS4gU28gaXMgZWFjaCBzaGFkZWQgcmVjdGFuZ2xlLiBJZiBhIHNoYWRlZCByZWN0YW5nbGUgaXMgYSBjbGFkZSB3aXRoIG9ubHkgb25lIHNwZWNpZXMgKHN1Y2ggYXMgd2l0aCB0aGUgR3JlZW4gV2FyYmxlciBGaW5jaCBvciB0aGUgVmVnZXRhcmlhbiBGaW5jaCksIGNvdW50IGl0IG9ubHkgb25jZS4=
Cg==[Qq]
[f]IE5pY2UhIEEgY2xhZGUgaXMgYSBncm91cCB0aGF0IGNvbnNpc3RzIG9mIGEgY29tbW9uIGFuY2VzdG9yIGFuZCBhbGwgb2YgaXRzIGRlc2NlbmRhbnRzLiBJbiB0aGUgdHJlZSBhYm92ZSwgdGhlcmUgYXJlIDExIGNsYWRlcy4gSWYgeW91IHdhbnQgdG8gZG91YmxlLWNoZWNrIHlvdXIgYW5zd2VyLCB1c2UgdGhlIGRpYWdyYW0gYmVsb3cu
Cg==Cg==[Qq]
[q json=”true” multiple_choice=”true” unit=”7.Evolution” topic=”7.9.Phylogeny” dataset_id=”2019 AP Bio Dataset|1d25c97fa8cf30″ question_number=”380″] Which of the following is the shared derived trait that unites rabbits, monkeys, alligators, and chickens into a single clade?
[c]IEhhaXIgYW5kIGFuIGVnZyB3aXRoIGEgaGFyZCBzaGVsbC4=[Qq]
[f]IE5vLiBIYWlyIGlzIGEgc2hhcmVkIGRlcml2ZWQgdHJhaXQgb2YgbWFtbWFscyAoaW5jbHVkaW5nIHJhYmJpdHMgYW5kIG1vbmtleXMpLiBBbiBlZ2cgd2l0aCBhIGhhcmQgc2hlbGwgaXMgYSBzaGFyZWQgZGVyaXZlZCB0cmFpdCBvZiB0aGUgY2xhZGUgdGhhdCBpbmNsdWRlcyBhbGxpZ2F0b3JzIGFuZCBjaGlja2Vucy4gV2hhdCYjODIxNztzIHRoZSBzaGFyZWQgZGVyaXZlZCB0cmFpdCBvZiByYWJiaXRzLCBtb25rZXlzLCBhbGxpZ2F0b3JzLCA=YW5kIGNoaWNrZW5zPw==[Qq]
[c]IEhhdmluZyBmb3VyIGxpbWJz[Qq]
[f]IE5vLiBIYXZpbmcgZm91ciBsaW1icyBpcyBhbiBhbmNlc3RyYWwgdHJhaXQgb2YgcmFiYml0cywgbW9ua2V5cywgYWxsaWdhdG9ycywgYW5kIGNoaWNrZW5zLiBGaW5kIGEgc2hhcmVkIGRlcml2ZWQgdHJhaXQgdGhhdCB1bml0ZXMgcmFiYml0cywgbW9ua2V5cywgYWxsaWdhdG9ycywgYW5kIGNoaWNrZW5zIHdpdGhpbiBhIHNpbmdsZSBjbGFkZS4=[Qq]
[c]IEFuIGFtbmlv dGljIGVnZw==[Qq]
[f]IE5pY2UuIEFuIGFtbmlvdGljIGVnZyBpcyBhIHNoYXJlZCBkZXJpdmVkIHRyYWl0IHRoYXQgdW5pdGVzIHJhYmJpdHMsIG1vbmtleXMsIGFsbGlnYXRvcnMsIGFuZCBjaGlja2VucyB3aXRoaW4gYSBzaW5nbGUgY2xhZGUu[Qq]
[c]IEEgdmVydGVicmFsIGNvbHVtbi4=[Qq]
[f]IE5vLiBIYXZpbmcgYSB2ZXJ0ZWJyYWwgY29sdW1uIGlzIHRoZSBzaGFyZWQgZGVyaXZlZCB0cmFpdCBvZiBhbGwgdmVydGVicmF0ZXMuIEl0JiM4MjE3O3MgYW4gYW5jZXN0cmFsIHRyYWl0IG9mIHJhYmJpdHMsIG1vbmtleXMsIGFsbGlnYXRvcnMsIGFuZCBjaGlja2Vucy4gRmluZCBhIHNoYXJlZCBkZXJpdmVkIHRyYWl0IHRoYXQgdW5pdGVzIHJhYmJpdHMsIG1vbmtleXMsIGFsbGlnYXRvcnMsIGFuZCBjaGlja2VucyB3aXRoaW4gYSBzaW5nbGUgY2xhZGUu
Cg==[Qq]
[q json=”true” multiple_choice=”true” unit=”7.Evolution” topic=”7.9.Phylogeny” dataset_id=”2019 AP Bio Dataset|1d2485dd340330″ question_number=”381″] The table below shows the number of amino acid differences in cytochrome c (a protein that’s part of the electron transport chain) in 5 different species. All versions of cytochrome c consist of 104 amino acids.
By correlating the data in the table below with data in the fossil record, it’s been determined that chickens and penguins had a common ancestor about 70 million years ago. Using that data, a scientist infers that the lineage leading to snakes split from the lineage leading to birds about 360 million years ago. This inference is based on the idea that
[c]IGJpb2xvZ2ljYWwgbW9sZWN1bGVzIHdpdGggaW1wb3J0YW50IGZ1bmN0aW9ucyByZW1haW4gc3RhYmxlIG92ZXIgdGltZS4=[Qq]
[f]IE5vLiBBbGwgb2YgdGhlIG9yZ2FuaXNtcyBhYm92ZSBoYXZlIHRoZSBjeXRvY2hyb21lIGMgcHJvdGVpbi4gWWV0IHRoZXJlIGFyZSBzaWduaWZpY2FudCBkaWZmZXJlbmNlcyBpbiB0aGUgYW1pbm8gYWNpZHMgbWFraW5nIHVwIGN5dG9jaHJvbWUgYy4gVGhhdCBtZWFucyB0aGF0IG92ZXIgdGltZSwgdGhlIG1vbGVjdWxlIGhhcyBldm9sdmVkIChhbmQgbm90IHJlbWFpbmVkIHN0YWJsZSku[Qq]
[c]IG11dGF0aW9ucyBhY2N1bXVsYXRlIGF0IG Egcm91Z2hseSBjb25zdGFudCByYXRlLg==[Qq]
[f]IEZhYnVsb3VzLiBUaGUgZGF0YSBhYm92ZSBpcyB0aGUgYmFzaXMgZm9yIHVzaW5nIGN5dG9jaHJvbWUgYyBhcyBhIG1vbGVjdWxhciBjbG9jay4gVGhhdCBpZGVhLCBpbiB0dXJuLCBpcyBiYXNlZCBvbiB0aGF0IG11dGF0aW9ucyBhY2N1bXVsYXRlIGF0IGEgY29uc3RhbnQgcmF0ZS4=[Qq]
[c]IHNoYXJlZCBkZXJpdmVkIHRyYWl0cyBhcmlzZSBxdWlja2x5IG92ZXIgdGhlIGNvdXJzZSBvZiBldm9sdXRpb24u[Qq]
[f]IE5vLiBTaGFyZWQgZGVyaXZlZCB0cmFpdHMgYXJlIHRyYWl0cyB0aGF0IGFyZSBmb3VuZCBpbiBhIGNvbW1vbiBhbmNlc3RvciBhbmQgYWxsIG9mIGl0cyBkZXNjZW5kYW50cywgYW5kIHdoaWNoIGRlZmluZSBhIGNsYWRlLg==
Cg==VGhpcyBxdWVzdGlvbiBpcyBhYm91dCB0aGUgaWRlYSBvZiBhIG1vbGVjdWxlIGNsb2NrLCB3aGljaCBjb3JyZWxhdGVzIG1vbGVjdWxhciBkaWZmZXJlbmNlcyB3aXRoIHRoZSB0aW1lIG9mIGRpdmVyZ2VuY2UuIE5vdGUgdGhlIHNoYXBlIG9mIHRoZSBsaW5lIGluIHRoZSBjeXRvY2hyb21lIGMgbW9sZWN1bGFyIGNsb2NrIGJlbG93LiBJcyBpdCBjaGFuZ2luZyBvdmVyIHRpbWUsIG9yIGRvZXMgaXQgYXBwZWFyIHRvIGJlIGNvbnN0YW50Pw==
Cg==[Qq]
[c]IE1vcnBob2xvZ2ljYWwgc2ltaWxhcml0aWVzIHJlZmxlY3QgZXZvbHV0aW9uYXJ5IGhvbW9sb2d5LiBNb2xlY3VsYXIgc2ltaWxhcml0aWVzIGV2b2x2ZSBmcm9tIGNvbnZlcmdlbnQgZXZvbHV0aW9uLg==[Qq]
[f]IE5vLCBmb3IgdHdvIHJlYXNvbnMuIEZpcnN0LCBtb2xlY3VsYXIgc2ltaWxhcml0aWVzIHJlZmxlY3QgaG9tb2xvZ3kgbW9yZSBvZnRlbiB0aGFuIG1vcnBob2xvZ3ksIHdoaWNoIGNhbiByZXN1bHQgZnJvbSBjb252ZXJnZW50IGV2b2x1dGlvbi4gU2Vjb25kLCB0aGF0JiM4MjE3O3Mgbm90IHdoYXQgdGhlIHF1ZXN0aW9uIGlzIGFib3V0Lg==
Cg==VGhpcyBxdWVzdGlvbiBpcyBhYm91dCB0aGUgaWRlYSBvZiBhIG1vbGVjdWxlIGNsb2NrLCB3aGljaCBjb3JyZWxhdGVzIG1vbGVjdWxhciBkaWZmZXJlbmNlcyB3aXRoIHRoZSB0aW1lIG9mIGRpdmVyZ2VuY2UuIE5vdGUgdGhlIHNoYXBlIG9mIHRoZSBsaW5lIGluIHRoZSBjeXRvY2hyb21lIGMgbW9sZWN1bGFyIGNsb2NrIGJlbG93LiBJcyBpdCBjaGFuZ2luZyBvdmVyIHRpbWUsIG9yIGRvZXMgaXQgYXBwZWFyIHRvIGJlIGNvbnN0YW50Pw==
Cg==[Qq]
[q json=”true” multiple_choice=”true” unit=”7.Evolution” topic=”7.9.Phylogeny” dataset_id=”2019 AP Bio Dataset|1d22939b436b30″ question_number=”382″] The table below shows the number of amino acid differences in cytochrome c (a protein that’s part of the electron transport chain) in 5 different species. In all of these species, cytochrome c consists of 104 amino acids.
By correlating the data in the table below with data in the fossil record, it’s been determined that chickens and penguins had a common ancestor about 70 million years ago. Using that data, a scientist is attempting to determine when the lineage leading to chickens split from the lineage leading to yeast. From the choices below, what’s the most reasonable estimate?
[c]IDQ2IG1pbGxpb24geWVhcnM=[Qq]
[f]IE5vLiA0NiBpcyB0aGUgbnVtYmVyIG9mIGFtaW5vIGFjaWQgZGlmZmVyZW5jZXMgaW4gY3l0b2Nocm9tZSBjIGJldHdlZW4gY2hpY2tlbnMgYW5kIHllYXN0LiBUbyBkZXRlcm1pbmUgdGhlIHRpbWUgb2YgZGl2ZXJnZW5jZSwgeW91IGhhdmUgdG8gZG8gc29tZSBhbGdlYnJhLiBJZiBpdCB0b29rIDcwIG1pbGxpb24geWVhcnMgZm9yIDMgZGlmZmVyZW5jZXMgdG8gYWNjdW11bGF0ZSwgaG93IGxvbmcgd291bGQgaXQgdGFrZSBmb3IgNDYgZGlmZmVyZW5jZXMgdG8gYWNjdW11bGF0ZT8=[Qq]
[c]IDEzOCBtaWxsaW9uIHllYXJz[Qq]
[f]IE5vLiBJdCBsb29rcyBsaWtlIHlvdSBtdWx0aXBsaWVkIHRoZSBudW1iZXIgb2YgY3l0b2Nocm9tZSBjIGFtaW5vIGFjaWQgZGlmZmVyZW5jZXMgYmV0d2VlbiB5ZWFzdCBhbmQgY2hpY2tlbnMgKDQ2KSB3aXRoIHRoZSBudW1iZXIgb2YgYW1pbm8gYWNpZCBkaWZmZXJlbmNlcyBiZXR3ZWVuIGNoaWNrZW5zIGFuZCBwZW5ndWlucyAoMyku
Cg==VG8gZGV0ZXJtaW5lIHRoZSB0aW1lIG9mIGRpdmVyZ2VuY2UsIHlvdSBoYXZlIHRvIGRvIHNvbWUgYWxnZWJyYS4gSWYgaXQgdG9vayA3MCBtaWxsaW9uIHllYXJzIGZvciAzIGRpZmZlcmVuY2VzIHRvIGFjY3VtdWxhdGUsIGhvdyBsb25nIHdvdWxkIGl0IHRha2UgZm9yIDQ2IGRpZmZlcmVuY2VzIHRvIGFjY3VtdWxhdGU/[Qq]
[c]IDMuMiBiaWxsaW9uIHllYXJz[Qq]
[f]IE5vLiBJdCBsb29rcyBsaWtlIHlvdSBtdWx0aXBsaWVkIHRoZSBudW1iZXIgb2YgY3l0b2Nocm9tZSBjIGFtaW5vIGFjaWQgZGlmZmVyZW5jZXMgYmV0d2VlbiB5ZWFzdCBhbmQgY2hpY2tlbnMgKDQ2KSBieSB0aGUgbnVtYmVyIG9mIHllYXJzIGl0IHRvb2sgdGhvc2UgZGlmZmVyZW5jZXMgdG8gYWNjdW11bGF0ZSAoNzAgbWlsbGlvbikgdG8gZ2V0IDMuMiBiaWxsaW9uLg==
Cg==VG8gZGV0ZXJtaW5lIHRoZSB0aW1lIG9mIGRpdmVyZ2VuY2UsIHlvdSBoYXZlIHRvIGRvIHNvbWUgYWxnZWJyYS4gSWYgaXQgdG9vayA3MCBtaWxsaW9uIHllYXJzIGZvciAzIGRpZmZlcmVuY2VzIHRvIGFjY3VtdWxhdGUsIGhvdyBsb25nIHdvdWxkIGl0IHRha2UgZm9yIDQ2IGRpZmZlcmVuY2VzIHRvIGFjY3VtdWxhdGU/[Qq]
[c]IDkyMCBtaWxsaW 9uIHllYXJzLg==[Qq]
[f]IEV4Y2VsbGVudC4gSWYgaXQgdG9vayA3MCBtaWxsaW9uIHllYXJzIGZvciAzIGRpZmZlcmVuY2VzIHRvIGFjY3VtdWxhdGUsIHRoZW4gaXQgd291bGQgdGFrZSA5MjAgbWlsbGlvbiB5ZWFycyBmb3IgNDYgZGlmZmVyZW5jZXMgdG8gYWNjdW11bGF0ZS4gTm90ZSB0aGF0IHdoaWxlIHRoaXMgaXMgdGhlIGNvcnJlY3QgYW5zd2VyIGJhc2VkIG9uIHRoZSBpbmZvcm1hdGlvbiB5b3Ugd2VyZSBnaXZlbiwgdGhlIGFjdHVhbCBkYXRlIG9mIGRpdmVyZ2VuY2UgYmV0d2VlbiB2ZXJ0ZWJyYXRlcyBhbmQgeWVhc3QgcHJvYmFibHkgZ29lcyBiYWNrIHRvIHRoZSBvcmlnaW4gb2YgZXVrYXJ5b3Rlcywgd2hpY2ggaXMgdGhvdWdodCB0byBoYXZlIG9jY3VycmVkIGJldHdlZW4gMS42IGJpbGxpb24gdG8gMiBiaWxsaW9uIHllYXJzIGFnby4=
Cg==[Qq]
[q json=”true” xyz=”2″ multiple_choice=”true” dataset_id=”2019 AP Bio Dataset|21182e588c445a” question_number=”383″ unit=”7.Evolution” topic=”7.9.Phylogeny”] Which of the following statements is consistent with the phylogenetic tree below?
[c]IExhbXByZXlzIGFuZCBtYW1tYWxzIGFyZSBub3QgcmVsYXRlZC4=[Qq]
[f]IE5vLiBKdXN0IGZvbGxvdyB0aGUgdHJlZSBiYWNrIGluIHRpbWUgKHRvIHRoZSBsZWZ0KSBhbmQgeW91JiM4MjE3O2xsIHNlZSB0aGF0IGxhbXByZXlzIGFuZCBtYW1tYWxzIGFyZSByZWxhdGVkLCBib3RoIGRlc2NlbmRlZCBmcm9tIGEgY29tbW9uIGFuY2VzdG9yLg==[Qq]
[c]IENhcnRpbGFnaW5vdXMgZmlzaCBhcmUgdGhlIGFuY2VzdG9ycyBvZiBhbXBoaWJpYW5zLg==[Qq]
[f]IE5vLiBBbGwgeW91IGNhbiBpbmZlciBmcm9tIHRoaXMgcGh5bG9nZW55IGlzIHRoYXQgMSkgY2FydGlsYWdpbm91cyBmaXNoIGVtZXJnZWQgZWFybGllciB0aGFuIGFtcGhpYmlhbnMgZGlkLCBhbmQgMikgY2FydGlsYWdpbm91cyBmaXNoIGFuZCBhbXBoaWJpYW5zIGhhdmUgYSBjb21tb24gYW5jZXN0b3IuIERvbiYjODIxNzt0IGNvbmZ1c2UgJiM4MjIwO2VhcmxpZXIgZW1lcmdlbmNlJiM4MjIxOyB3aXRoICYjODIyMDtiZWluZyBhbmNlc3RyYWwgdG8uJiM4MjIxOyBUbyByZXN0YXRlIHRoaXMgaW4gdGVybXMgYSBiaXQgY2xvc2VyIHRvIHVzIGh1bWFucywgcmVtZW1iZXIgdGhhdCBodW1hbnMgYW5kIGNoaW1wcyBzaGFyZSBhIGNvbW1vbiBhbmNlc3RvciAoYWJvdXQgNiBtaWxsaW9uIHllYXJzIGFnbykuIEJ1dCBjaGltcHMgYXJlIG5vdCBhbmNlc3RyYWwgdG8gaHVtYW5zLiBXZSYjODIxNzt2ZSBib3RoIGJlZW4gc2VwYXJhdGVseSBldm9sdmluZyBzaW5jZSBvdXIgc3BsaXQgZnJvbSB0aGUgY29tbW9uIGFuY2VzdG9yLg==[Qq]
[c]IEJpcmRzIGFyZSBtb3JlIGNsb3NlbHkgcmVsYXRlZCB0byByZXB0aWxlcyB0aGFuIHRvIG1hbW1hbHMu[Qq]
[f]IE5vLiBUaGlzIHBoeWxvZ2VueSBzdWdnZXN0cyB0aGF0IGJpcmRzIGFuZCBtYW1tYWxzIHNoYXJlIGEgbW9yZSByZWNlbnQgY29tbW9uIGFuY2VzdG9yIHRoYW4gZG8gYmlyZHMgYW5kIHJlcHRpbGVzLg==[Qq]
[c]IEFtcGhpYmlhbnMsIHJlcHRpbGVzLCBiaXJkcywgYW5k IG1hbW1hbHMgc2hhcmUgYSBjb21tb24gYW5jZXN0b3Iu[Qq]
[f]IEZhYnVsb3VzLiBUaGlzIHBoeWxvZ2VueSBzdWdnZXN0cyB0aGF0IGFtcGhpYmlhbnMsIHJlcHRpbGVzLCBiaXJkcywgYW5kIG1hbW1hbHMgc2hhcmUgYSBjb21tb24gYW5jZXN0b3Iu[Qq]
[q json=”true” xyz=”2″ multiple_choice=”true” dataset_id=”2019 AP Bio Dataset|211542f5a3605a” question_number=”384″ unit=”7.Evolution” topic=”7.9.Phylogeny”] Cytochrome c is a protein that’s part of the mitochondrial electron transport chain. By comparing amino acid similarities and differences in cytochrome c among various organisms, one can make inferences about these species’ genetic relatedness.
The table below shows differences in cytochrome c amino acid sequences for nine species:
Based on the data above, what species should be in position 7 in the phylogenetic tree below?
[c]IHdoZWF0[Qq]
[f]IE5vLiBTdGFydCBieSBub3RpbmcgdGhhdCBob3JzZXMgYW5kIGRvbmtleXMgaGF2ZSBvbmx5IG9uZSBkaWZmZXJlbmNlLCBzdWdnZXN0aW5nIHRoYXQgdGhleSBoYXZlIHRoZSBtb3N0IHJlY2VudCBjb21tb24gYW5jZXN0b3IuIFRoYXQgcHV0cyB0aGVtIGludG8gcG9zaXRpb25zIDkgKGhvcnNlKSBhbmQgOChkb25rZXkpLiBCYXNlZCBvbiB0aGUgY2hhcnQgKGFuZCwgb2YgY291cnNlLCB3aGF0IHlvdSBrbm93IGFib3V0IGFuaW1hbHMpLCB3aGF0JiM4MjE3O3MgdGhlIG5leHQgY2xvc2VzdCByZWxhdGl2ZSBvZiB0aGUgaG9yc2UgYW5kIHRoZSBkb25rZXk/[Qq]
[c]IG1vdGg=[Qq]
[f]IE5vLiBTdGFydCBieSBub3RpbmcgdGhhdCBob3JzZXMgYW5kIGRvbmtleXMgaGF2ZSBvbmx5IG9uZSBkaWZmZXJlbmNlLCBzdWdnZXN0aW5nIHRoYXQgdGhleSBoYXZlIHRoZSBtb3N0IHJlY2VudCBjb21tb24gYW5jZXN0b3IuIFRoYXQgcHV0cyB0aGVtIGludG8gcG9zaXRpb25zIDkgKGhvcnNlKSBhbmQgOCAoZG9ua2V5KS4gQmFzZWQgb24gdGhlIGNoYXJ0IChhbmQsIG9mIGNvdXJzZSwgd2hhdCB5b3Uga25vdyBhYm91dCBhbmltYWxzKSwgd2hhdCYjODIxNztzIHRoZSBuZXh0IGNsb3Nlc3QgcmVsYXRpdmUgb2YgdGhlIGhvcnNlIGFuZCB0aGUgZG9ua2V5Pw==[Qq]
[c]IHdo YWxl[Qq]
[f]IEV4Y2VsbGVudCEgVGhlIGRvbmtleSBhbmQgaG9yc2UsIGJhc2VkIG9uIGN5dG9jaHJvbWUgYyAoYW5kLCBvZiBjb3Vyc2UsIHlvdXIgcHJpb3Iga25vd2xlZGdlKSBhcmUgdGhlIG1vc3QgY2xvc2VseSByZWxhdGVkIHNwZWNpZXMuIFRoZSBuZXh0IGlzIHRoZSB3aGFsZSAod2l0aCA1IGRpZmZlcmVuY2VzIGZyb20gdGhlIGhvcnNlLCBhbmQgNCBmcm9tIHRoZSBkb25rZXkpLg==[Qq]
[c]IHBlbmd1aW4=[Qq]
[f]IE5vLiBTdGFydCBieSBub3RpbmcgdGhhdCBob3JzZXMgYW5kIGRvbmtleXMgaGF2ZSBvbmx5IG9uZSBkaWZmZXJlbmNlLCBzdWdnZXN0aW5nIHRoYXQgdGhleSBoYXZlIHRoZSBtb3N0IHJlY2VudCBjb21tb24gYW5jZXN0b3IuIFRoYXQgcHV0cyB0aGVtIGludG8gcG9zaXRpb25zIDkgKGhvcnNlKSBhbmQgOCAoZG9ua2V5KS4gQmFzZWQgb24gdGhlIGNoYXJ0IChhbmQsIG9mIGNvdXJzZSwgd2hhdCB5b3Uga25vdyBhYm91dCBhbmltYWxzKSwgd2hhdCYjODIxNztzIHRoZSBuZXh0IGNsb3Nlc3QgcmVsYXRpdmUgb2YgdGhlIGhvcnNlIGFuZCB0aGUgZG9ua2V5Pw==[Qq]
[c]IHNuYWtl[Qq]
[f]IE5vLiBTdGFydCBieSBub3RpbmcgdGhhdCBob3JzZXMgYW5kIGRvbmtleXMgaGF2ZSBvbmx5IG9uZSBkaWZmZXJlbmNlLCBzdWdnZXN0aW5nIHRoYXQgdGhleSBoYXZlIHRoZSBtb3N0IHJlY2VudCBjb21tb24gYW5jZXN0b3IuIFRoYXQgcHV0cyB0aGVtIGludG8gcG9zaXRpb25zIDkgKGhvcnNlKSBhbmQgOCAoZG9ua2V5KS4gQmFzZWQgb24gdGhlIGNoYXJ0IChhbmQsIG9mIGNvdXJzZSwgd2hhdCB5b3Uga25vdyBhYm91dCBhbmltYWxzKSwgd2hhdCYjODIxNztzIHRoZSBuZXh0IGNsb3Nlc3QgcmVsYXRpdmUgb2YgdGhlIGhvcnNlIGFuZCB0aGUgZG9ua2V5Pw==[Qq]
[q json=”true” xyz=”2″ multiple_choice=”true” dataset_id=”2019 AP Bio Dataset|211259e6c6605a” question_number=”385″ unit=”7.Evolution” topic=”7.9.Phylogeny”] The human phylogenetic tree below represents the relationship between different groups.
According to the tree, unique Neanderthal DNA is least likely to be found in which group?
[c]IEhhbg==[Qq]
[f]IE5vLiBOb3RlIHRoZSB2ZXJ0aWNhbCBsaW5lLCB3aGljaCBpbmRpY2F0ZXMgaW50ZXJicmVlZGluZyBiZXR3ZWVuIGdyb3Vwcy4gVGhlIE5lYW5kZXJ0aGFscyBpbnRlcmJyZWQgd2l0aCB0aGUgY29tbW9uIGFuY2VzdG9ycyBvZiB0aGUgSGFuLCBNZWxhbmVzaWFuLCBhbmQgRnJlbmNoLCBhbmQgdGhhdCB3b3VsZCBoYXZlIHByb3ZpZGVkIGEgd2F5IGZvciBOZWFuZGVydGhhbCBnZW5lcyB0byBmbG93IGludG8gdGhlIEhhbiBwb3B1bGF0aW9uLg==[Qq]
[c]IEFmcm ljYW4=[Qq]
[f]IFRoYXQmIzgyMTc7cyByaWdodC4gVGhlIE5lYW5kZXJ0aGFscyBldm9sdmVkIGFmdGVyIHRoZWlyIHNwbGl0IGZyb20gdGhlIGxhc3QgY29tbW9uIGFuY2VzdG9yIHdpdGggdGhlIGdyb3VwIHRoYXQmIzgyMTc7cyBkZXNpZ25hdGVkIGFzICYjODIyMDtBZnJpY2FuLiYjODIyMTsgRnVydGhlcm1vcmUsIHRoZXJlIHdhcyBubyBpbnRlcmJyZWVkaW5nIGJldHdlZW4gdGhlc2UgdHdvIGdyb3Vwcy4gQXMgYSByZXN1bHQsIGFueSB1bmlxdWUgTmVhbmRlcnRoYWwgRE5BIHNob3VsZCBub3QgYmUgcHJlc2VudCBpbiBBZnJpY2Fucy4=[Qq]
[c]IE1lbGFuZXNpYW4=[Qq]
[f]IE5vLiBOb3RlIHRoZSB2ZXJ0aWNhbCBsaW5lLCB3aGljaCBpbmRpY2F0ZXMgaW50ZXJicmVlZGluZyBiZXR3ZWVuIGdyb3Vwcy4gVGhlIE5lYW5kZXJ0aGFscyBpbnRlcmJyZWQgd2l0aCB0aGUgY29tbW9uIGFuY2VzdG9ycyBvZiB0aGUgSGFuLCBNZWxhbmVzaWFuLCBhbmQgRnJlbmNoLCBhbmQgdGhhdCB3b3VsZCBoYXZlIHByb3ZpZGVkIGEgd2F5IGZvciBOZWFuZGVydGhhbCBnZW5lcyB0byBmbG93IGludG8gdGhlIE1lbGFuZXNpYW4gcG9wdWxhdGlvbi4=[Qq]
[c]IERlbmlzb3Zh[Qq]
[f]IE5vLiBOb3RlIHRoZSB2ZXJ0aWNhbCBsaW5lcywgd2hpY2ggaW5kaWNhdGUgaW50ZXJicmVlZGluZyB3aXRoaW4gZ3JvdXBzLiBUaGUgTmVhbmRlcnRoYWxzIGludGVyYnJlZCB3aXRoIHRoZSBjb21tb24gYW5jZXN0b3JzIG9mIHRoZSBIYW4sIE1lbGFuZXNpYW4sIGFuZCBGcmVuY2gsIGFuZCB0aGF0IHdvdWxkIGhhdmUgcHJvdmlkZWQgYSB3YXkgZm9yIE5lYW5kZXJ0aGFsIGdlbmVzIHRvIGZsb3cgaW50byB0aGUgTWVsYW5lc2lhbiBwb3B1bGF0aW9uLiBUaGVuLCB0aGUgTWVsYW5lc2lhbnMgaW50ZXJicmVkIHdpdGggdGhlIERlbmlzb3ZhLCBwcm92aWRpbmcgYSB3YXkgZm9yIE5lYW5kZXJ0aGFsIGdlbmVzIHRvIGZsb3cgZnJvbSB0aGUgTWVsYW5lc2lhbnMgdG8gdGhlIERlbmlzb3ZhLg==[Qq]
[q json=”true” xyz=”2″ multiple_choice=”true” dataset_id=”2019 AP Bio Dataset|210f732bf5445a” question_number=”386″ unit=”7.Evolution” topic=”7.9.Phylogeny”] Cytochrome c is a protein found in the mitochondrial electron transport chain. The amino acid sequence of this protein varies between species and can be used to determine evolutionary relationships. Scientists studied the number of differences in the amino acid sequences in cytochrome c between three species of chordate (A, B, and C). The results of the study are shown in the table below.
| Species B | Species C | |
| Species A | 11 | 3 |
| Species B | 0 | 10 |
Which of the following phylogenetic trees is most consistent with the results of the study?
[c]IG E=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIGN5dG9jaHJvbWUgYyBkYXRhIHRlbGxzIHlvdSB0aGF0IHNwZWNpZXMgYSBhbmQgYyBhcmUgdGhlIG1vc3QgY2xvc2VseSByZWxhdGVkIChzaG93biBieSB0aGUgZmFjdCB0aGF0IHRoZWlyIGN5dG9jaHJvbWUgYyBzZXF1ZW5jZXMgZGlmZmVyIGJ5IG9ubHkgdGhyZWUgYW1pbm8gYWNpZHMpLiBGcm9tIHRoaXMsIHlvdSBjYW4gY29uY2x1ZGUgdGhhdCB0aGV5IHNoYXJlIGEgcmVjZW50IGNvbW1vbiBhbmNlc3RvciBhbmQgdGhhdCBib3RoIGFyZSBtb3JlIGRpc3RhbnRseSByZWxhdGVkIHRvIHNwZWNpZXMgYi4=[Qq]
[c]IGI=[Qq]
[f]IE5vLiBMb29rIGF0IHRoZSBkYXRhIHNldC4gV2hpY2ggb2YgdGhlIHR3byBzcGVjaWVzIGFyZSBtb3N0IGNsb3NlbHkgcmVsYXRlZD8gSXQmIzgyMTc7cyB0aGUgdHdvIHdpdGggdGhlIGxlYXN0IG51bWJlciBvZiBkaWZmZXJlbmNlcyBpbiB0aGVpciBjeXRvY2hyb21lIGMgc2VxdWVuY2UuIFVzZSB5b3VyIGFuc3dlciB0byB0aGF0IHF1ZXN0aW9uIGFzIGEgd2F5IHRvIGNob29zZSB0aGUgY29ycmVjdCBwaHlsb2dlbnku[Qq]
[c]IGM=[Qq]
[f]IE5vLiBMb29rIGF0IHRoZSBkYXRhIHNldC4gV2hpY2ggb2YgdGhlIHR3byBzcGVjaWVzIGFyZSBtb3N0IGNsb3NlbHkgcmVsYXRlZD8gSXQmIzgyMTc7cyB0aGUgdHdvIHdpdGggdGhlIGxlYXN0IG51bWJlciBvZiBkaWZmZXJlbmNlcyBpbiB0aGVpciBjeXRvY2hyb21lIGMgc2VxdWVuY2UuIFVzZSB5b3VyIGFuc3dlciB0byB0aGF0IHF1ZXN0aW9uIGFzIGEgd2F5IHRvIGNob29zZSB0aGUgY29ycmVjdCBwaHlsb2dlbnku[Qq]
[c]IGQ=[Qq]
[f]IE5vLiBUaGUgdHJlZSB5b3UmIzgyMTc7dmUgY2hvc2VuIHN1Z2dlc3RzIHRoYXQgc3BlY2llcyBhIGFuZCBiIGFyZSB0aGUgbW9zdCBjbG9zZWx5IHJlbGF0ZWQuIE5vdyBsb29rIGFnYWluIGF0IHRoZSBkYXRhIHNldC4gVGhlIHR3byBtb3N0IGNsb3NlbHkgcmVsYXRlZCBhcmUgdGhlIG9uZXMgd2l0aCB0aGUgbGVhc3QgbnVtYmVyIG9mIGRpZmZlcmVuY2VzIGluIHRoZWlyIGN5dG9jaHJvbWUgYyBzZXF1ZW5jZS4gVXNlIHRoYXQgdG8gY2hvb3NlIHRoZSBjb3JyZWN0IHBoeWxvZ2VueSB0aGUgbmV4dCB0aW1lIHlvdSBzZWUgdGhpcyBxdWVzdGlvbi4=[Qq]
[/qwiz]
What’s Next?
Please proceed to the next tutorial: Origin of Life
