1. Watch this video
2a. Read this: Some additional useful concepts and terms
- Eukaryotic cells have vast amounts of DNA. Stretched out end to end, the DNA in a human cell would be several meters in length. To package the DNA into the nucleus, the DNA is wound around structures called nucleosomes, which are composed of histone proteins.
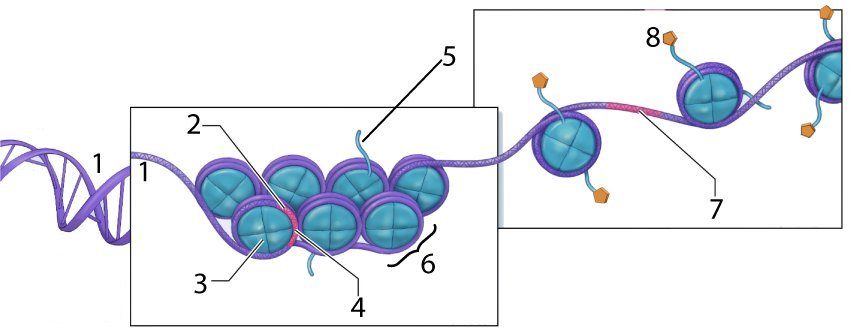
The DNA is shown at “1.” At “2” we have a gene, but it’s wrapped up so tightly that it’s unavailable for transcription (with “4” representing the untranscribable DNA). Number “6” is a nucleosome, a “bead” consisting of several linked histone proteins (one of which is indicated by “3”). - Euchromatin: loosely packed (often acetylated) DNA that is transcribed into RNA and translated into protein.
- Heterochromatin: tightly packed (often methylated) DNA that is NOT transcribed
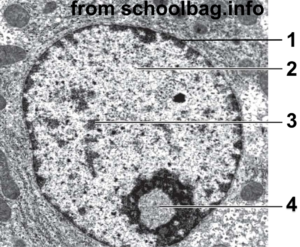
1. Nuclear membrane. 2. Euchromatin 3. Heterochromatin 4. Nucleolus
2b. Study this summary
Why Gene Regulation in Eukaryotes is Complex (especially when compared to prokaryotic gene regulation)
- Size: Eukaryotic genomes are much larger than prokaryotic genomes (4 billion base pairs in the human haploid genome, vs. 3 million base pairs in E. coli).
- Control of Tissue-Specific Gene Expression: Multicellular eukaryotes (e.g., humans, lizards, jellyfish) have trillions of cells with the same DNA, but each tissue expresses specific genes based on its function (e.g., muscle vs. skin cells). How do cells know which genes to express?
- Coding vs. Non-Coding DNA: Eukaryotic genes are not contiguous but interrupted by non-coding DNA stretches called introns. How do cells “know” to express the coding DNA, but not the intron?
Mechanisms of Eukaryotic Gene Regulation:
- Epigenetic modifications of DNA.
- Changes in DNA expression through reversible chemical modifications without altering the DNA sequence.
- Methylation: Adds a methyl group to DNA, silencing gene transcription.
- Acetylation: Loosens DNA-histone complexes, enabling transcription.
- Drives tissue differentiation. For example, skin cells and muscle cells have the same genes, but express different proteins because of epigenetic differences.
- Some epigenetic changes can be passed to future generations (intergenerational transmission of epigenetic modification).
- Transcription Regulation:
- Involves regulatory DNA sequences and regulatory proteins:
- Promoters: Sites where RNA polymerase binds.
- Enhancers: Increase the likelihood of transcription.
- Activators, DNA-bending proteins, and transcription factors work together to enable RNA polymerase to transcribe genes.
- Involves regulatory DNA sequences and regulatory proteins:
- Coordination of Gene Expression:
- Different tissues can share common regulatory sequences.
- Example: Testosterone regulates gene expression in both neck (mane) and muscle tissues in male lions, causing tissue-specific effects.
RNA Processing in Eukaryotes:
- Introns and Exons:
- Introns: Non-coding sequences removed during RNA splicing.
- Exons: Coding sequences spliced together to form mRNA for translation.
- Alternative Splicing:
- Allows exons to be combined in various ways, producing multiple protein variants from one gene.
- Increases phenotypic variation.
- Post-Transcriptional Modifications in mRNA:
- 5′ GTP Cap:
- Protects mRNA from degradation.
- Aids in nuclear export and ribosome binding.
- 3′ Poly-A Tail:
- Enhances mRNA stability and delays breakdown.
- 5′ GTP Cap:
Role of Small RNAs:
- MicroRNAs (miRNAs):
- Regulate gene expression post-transcriptionally.
- Form RNA silencing complexes that either pause translation or degrade mRNA so that it can no longer be translated.
- Result: Altered gene expression without modifying DNA.
Why This Matters:
- Eukaryotic Gene Regulation:
- Ensures genes are expressed in the right place, at the right time, and in response to environmental or developmental signals.
- Essential for understanding processes like development, tissue specialization, and cellular responses.
3. Master these flashcards
[qdeck style=”width: 650px !important; min-height: 400px !important;” bold_text=”false” qrecord_id=”sciencemusicvideosMeister1961-Eukaryotic Gene Regulation, APBVP”]
[h] Eukaryotic Gene Regulation
[q] Why is gene regulation in eukaryotes more complex than in prokaryotes?
[a]
– Eukaryotic genomes are much larger (e.g., 4 billion base pairs in humans vs. 3 million in E. coli).
– Multicellular eukaryotes have trillions of cells with the same DNA, but different tissues express specific genes based on their function.
– Eukaryotic genes are interrupted by non-coding introns, requiring cells to selectively express coding DNA (exons).
[q] What are introns and exons?
[a]
– Introns: Non-coding sequences removed during RNA splicing.
– Exons: Coding sequences spliced together to form mRNA for translation.
[q] What are two types of epigenetic modifications?
[a]
1. Methylation: Adds methyl groups to DNA, silencing transcription.
2. Acetylation: Loosens DNA-histone complexes, enabling transcription.
[q] How do epigenetic modifications affect gene expression?
[a]
- Controls whether genes are expressed or not expressed.
- Drive tissue differentiation (e.g., skin vs. muscle cells)
- Some changes are reversible.
- Some changes can be passed to future generations.
[q] What is the role of promoters and enhancers in transcription regulation?
[a]
– Promoters: Sites where RNA polymerase binds to initiate transcription.
– Enhancers: Increase the likelihood of transcription by interacting with activators and other proteins.
[q] How is gene expression coordinated across different tissues?
[a]
– Shared regulatory sequences enable coordination.
– Example: Testosterone regulates the expression of different genes in the neck (mane) and muscle tissues in male lions, causing neck tissue to express more hair-related genes, and muscle tissue to enhance muscle growth.
[q] What is the role of small RNAs like microRNAs (miRNAs) in gene regulation?
[a]
– Regulate gene expression post-transcriptionally.
– Form RNA silencing complexes that either pause translation or degrade mRNA.
– Alter gene expression without modifying DNA.
[q] Why is eukaryotic gene regulation important?
[a]
– Ensures genes are expressed in the right place, at the right time, and in response to signals.
– Crucial for processes like development, tissue specialization, and cellular responses.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|18d2436940d10″ question_number=”220″ topic=”6.3.Transcription_and_RNA_Processing”] What is pre-mRNA? Describe some of the post-transcriptional modification that has to happen to pre-mRNA in eukaryotes before it can be translated into protein.
[a] In eukaryotic cells, pre-mRNA (2) is what’s transcribed from a protein-coding gene. Before it can be translated into protein, pre-mRNA is processed with addition of a 5’ GTP cap (“g”), a 3’ poly-A tail (j), excision of introns (“f”) and splicing together of exons.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” topic=”6.3.Transcription_and_RNA_Processing” dataset_id=”AP_Bio_Flashcards_2022|173d636aaee002″ question_number=”221″] What is the function of the 5′ GTP cap and the 3′ poly-A-tail that’s added to mRNA during eukaryotic RNA processing?
[a]
The 5’ GTP cap (“g”) protects the mRNA from breakdown by enzymes in the cytoplasm, and also assists the mRNA in leaving the nucleus and binding with a ribosome. The 3’ poly-A tail (j) makes the mRNA more stable and delays its enzymatic breakdown in the cytoplasm.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” topic=”6.3.Transcription_and_RNA_Processing” dataset_id=”AP_Bio_Flashcards_2022|ac507a8247bd” question_number=”223″]Explain how organization of eukaryotic genetic material into introns and exons can increase phenotypic variation.
[a] Through alternative splicing, exons can be spliced together in alternative ways allowing for the production of multiple protein versions from the same mRNA transcript.
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|18b2d4c8bad10″ question_number=”225″ topic=”6.4.Translation”] How are transcription and translation different in prokaryotes and eukaryotes?
[a] In prokaryotes (1), transcription and translation are coupled: protein synthesis begins while transcription of mRNA is still taking place. In eukaryotes (2), the processes of transcription and translation are spatially and temporally separate: transcription occurs in the nucleus, and translation is in the cytoplasm.
In prokaryotes, a single mRNA can code for multiple proteins within a specific metabolic pathway; in eukaryotes, the mRNA codes for only a single polypeptide. In eukaryotes, pre-mRNA is modified before translation occurs (the details of which are covered in another card).
[q json=”true” yy=”4″ unit=”6.Gene_Expression_and_Regulation” dataset_id=”AP_Bio_Flashcards_2022|18658f7e1c110″ question_number=”233″ topic=”6.5.Regulation_of_Gene_Expression”] What are transcription factors?
[a]
Transcription factors (5) are proteins that bind to DNA at or near a gene’s promoter (1). Transcription factors play a regulatory role, enhancing or inhibiting gene expression by promoting or inhibiting the binding of RNA polymerase (6)
[x][restart]
[/qdeck]
4. Tackle these quizzes
4.1. DNA Packaging (Euchromatin and Heterochromatin)/ Epigenetic Modifications of DNA
[qwiz qrecord_id=”sciencemusicvideosMeister1961-DNA Packaging, Epigenetics, APBVP”]
[h]DNA packaging and Epigenetics
[i]
[q labels = “top”]
[l]Acetyl group
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l]DNA
[fx] No. Please try again.
[f*] Excellent!
[l]histone
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]histone tail
[fx] No. Please try again.
[f*] Excellent!
[l]nucleosome
[fx] No. Please try again.
[f*] Correct!
[l]Transcribable DNA
[fx] No. Please try again.
[f*] Excellent!
[l]Untranscribable DNA
[fx] No. Please try again.
[f*] Correct!
[q labels = “top”]
[l]euchromatin
[fx] No. Please try again.
[f*] Excellent!
[l]heterochromatin
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]nuclear membrane
[fx] No. Please try again.
[f*] Excellent!
[l]nucleolus
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[q labels = “top”]
[l]active/transcribed DNA
[fx] No. Please try again.
[f*] Correct!
[l]inactive/untranscribed DNA
[fx] No. Please try again.
[f*] Correct!
[l]ribosome assembly site
[fx] No. Please try again.
[f*] Great!
[l]RNA must cross this to be transcribed
[fx] No. Please try again.
[f*] Correct!
[q labels = “top”]
[l]Acetyl group
[fx] No. Please try again.
[f*] Good!
[l]histone
[fx] No. Please try again.
[f*] Excellent!
[l]histone tail
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]methyl group
[fx] No. Please try again.
[f*] Correct!
[l]Transcribable DNA
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]Untranscribable DNA
[fx] No, that’s not correct. Please try again.
[f*] Good!
[q labels = “top”]
[l]acetyl group
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]euchromatin
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l]heterochromatin
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]methyl group
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]RNA polymerase
[fx] No. Please try again.
[f*] Great!
[q]The differences between muscle tissue and eye-lens tissue are not about differences in the [hangman] in these tissues. That’s because almost all cells in a eukaryotic organism are genomically [hangman]. The difference, in other words, isn’t genetic, but [hangman].
[c]RE5B[Qq]
[c]ZXF1aXZhbGVudA==[Qq]
[c]ZXBpZ2VuZXRpYw==[Qq]
[q] In the diagram below, which number is pointing to a histone?
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IEV4Y2VsbGVudC4gTnVtYmVyICYjODIyMDszJiM4MjIxOyBpcyBwb2ludGluZyB0byBhIGhpc3RvbmUu[Qq]
[c]IDc=[Qq]
[f]IFlvdSYjODIxNztyZSBleHRyZW1lbHkgY2xvc2UuICYjODIyMDs2JiM4MjIxOyBpcyBhIGdyb3VwIG9mIGhpc3RvbmVzIG9yZ2FuaXplZCBpbnRvIGEgbnVjbGVvc29tZS4gQ2hvb3NlIGEgbnVtYmVyIHRoYXQmIzgyMTc7cyBqdXN0IHBvaW50aW5nIHRvIG9uZSBoaXN0b25lLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIaXN0b25lcyBtYWtlIHVwIG51Y2xlb3NvbWVzIChzaG93biBhdCAmIzgyMjA7Ni4mIzgyMjE7KS4gVGhlIGhpc3RvbmVzIGhhdmUgdGFpbHMgKHNob3duIGF0ICYjODIyMDs1JiM4MjIxOykgdGhhdCBjYW4gYmUgdXNlZCB0byBtb2RpZnkgdGhlIGRlbnNpdHkgb2YgRE5BIHBhY2thZ2luZy4=[Qq]
[q] In the diagram below, which number is showing DNA that’s available for transcription?
[textentry single_char=”true”]
[c]ID c=[Qq]
[f]IEV4Y2VsbGVudCEgJiM4MjIwOzcmIzgyMjE7IHNob3dzIEROQSB0aGF0JiM4MjE3O3MgYXZhaWxhYmxlIGZvciB0cmFuc2NyaXB0aW9uLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIEROQSAoaW5kaWNhdGVkIGJ5ICYjODIyMDsxJiM4MjIxOykgdGhhdCYjODIxNztzICYjODIyMDtyZWxheGVkIGVub3VnaCB0byBiZSBhY2Nlc3NlZCBieSBtb2xlY3VsZXMgbGlrZSBSTkEgcG9seW1lcmFzZSwgbWFraW5nIHRyYW5zY3JpcHRpb24gcG9zc2libGUu[Qq]
[q] In the diagram below, which number is showing an acetyl group?
[textentry single_char=”true”]
[c]ID g=[Qq]
[f]IEdvb2Qgd29yay4gJiM4MjIwOzgmIzgyMjE7IGlzIGFuIGFjZXR5bCBncm91cCwgYXNzb2NpYXRlZCB3aXRoIGhpc3RvbmUgbW9kaWZpY2F0aW9uIHRoYXQgYWxsb3dzIHRyYW5zY3JpcHRpb24u[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIHNvbWV0aGluZyB0aGF0JiM4MjE3O3MgYm9uZGVkIHdpdGggb25lIG9mIHRoZSBoaXN0b25lIHRhaWxzIChzaG93biBhdCAmIzgyMjA7NSYjODIyMTspLCBhbGxvd2luZyB0aGUgRE5BIHRvIHVuY29pbCBhbmQgYmVjb21lIGF2YWlsYWJsZSBmb3IgdHJhbnNjcmlwdGlvbi4=[Qq]
[q] In the diagram below, which LETTER is showing DNA that is expressed?
[textentry single_char=”true”]
[c]IE I=[Qq]
[f]IE5pY2Ugam9iLiBCIGlzIHNob3dpbmcgRE5BIHRoYXQgd291bGQgYmUgZXhwcmVzc2VkIChldWNocm9tYXRpbiku[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIGxldHRlciB0aGF0JiM4MjE3O3MgYXNzb2NpYXRlZCB3aXRoIGxvb3NlbHkgY29pbGVkIEROQS4=[Qq]
[q] In the diagram below, which number represents methyl groups?
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IE5pY2UuICYjODIyMDs0JiM4MjIxOyByZXByZXNlbnRzIG1ldGh5bCBncm91cHMu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIG51bWJlciBpbmRpY2F0aW5nIHNvbWV0aGluZyB0aGF0IGNoZW1pY2FsbHkgbW9kaWZpZXMgRE5BIGFuZCBoaXN0b25lIHByb3RlaW5zIGluIGEgd2F5IHRoYXQgY2F1c2VzIEROQSB0byBiZWNvbWUgdGlnaHRseSBwYWNrZWQgaGV0ZXJvY2hyb21hdGluLg==[Qq]
[q] In the diagram below, which number represents an acetyl group?
[textentry single_char=”true”]
[c]ID Y=[Qq]
[f]IE5pY2UuICYjODIyMDs2JiM4MjIxOyByZXByZXNlbnRzIGFjZXR5bCBncm91cHMuIEFjZXR5bCBncm91cHMgbW9kaWZ5IGhpc3RvbmVzIGluIGEgd2F5IHRoYXQgY2F1c2VzIEROQSB0byB1bmNvaWwsIG1ha2luZyBnZW5lcyBhdmFpbGFibGUgZm9yIHRyYW5zY3JpcHRpb24u[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBhY2V0eWwgZ3JvdXBzIG1vZGlmeSBoaXN0b25lJiM4MjE3O3MgaW4gYSB3YXkgdGhhdCBhbGxvd3MgRE5BIHRvIHVuY29pbCwgbWFraW5nIGdlbmVzIGF2YWlsYWJsZSBmb3IgdHJhbnNjcmlwdGlvbi4=[Qq]
[q] The functional group associated with silencing DNA is the [hangman] group.
[c]IG1ldGh5bA==[Qq]
[q] The functional group associated with activating DNA so that it can be transcribed is the [hangman] group.
[c]IGFjZXR5bA==[Qq]
[/qwiz]
4.2. Control of Gene Expression through Transcription
[qwiz random = “true” style=”width: 650px !important; min-height: 400px !important;” qrecord_id=”sciencemusicvideosMeister1961-Eukaryotic Control of Gene Expression through Transcription, APBVP”]
[h]Eukaryotic Control of Gene Expression through Transcription
[i]
[q] In the diagram below, which letter indicates intron DNA (don’t worry about superscripts)?
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IE5pY2Ugam9iLiBUaGUgaW50cm9uIEROQSAoYXQgJiM4MjIwO2UmIzgyMjE7KSBiZWNvbWVzIFJOQSBpbiB0aGUgcHJlLVJOQSwgYnV0IHRoZW4gY3V0cyBjdXQgb3V0IGFzIG1STkEgaXMgZm9ybWVkLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLsKgVGhlIGludHJvbiBETkEgY29kZXMgZm9yIFJOQSB0aGF0IGdldHMgY3V0IG91dCBvZiB0aGUgcHJlLVJOQSBiZWZvcmUgaXQgZ2V0cyBtYWRlIGludG8gbVJOQS4gJiM4MjIwO0YmIzgyMjE7IHJlcHJlc2VudHMgdHdvIFJOQSBzZWdtZW50cyB0aGF0IGhhdmUgYmVlbiBjdXQgb3V0JiM4MjMwO3NvIGp1c3Qgd29yayB1cCBmcm9tIHRoZXJlIGFuZCB5b3UmIzgyMTc7bGwgaGF2ZSB0aGUgYW5zd2VyLg==[Qq]
[q] In the diagram below, pre-mRNA (still with introns) is represented by the number
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEF3ZXNvbWUuICYjODIyMDsyJiM4MjIxOyBpcyBwcmUtbVJOQS4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBTcGxpY2VkIG91dCBSTkEgaW50cm9ucyBjYW4gYmUgc2VlbiBhdCAmIzgyMjA7Zi4mIzgyMjE7IA==TG9vayBmb3IgUk5BIHRoYXQgc3RpbGwgY29udGFpbnMgaW50cm9ucy7CoA==wqA=[Qq]
[q] In the diagram below, messenger RNA is represented by the number
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IEdvb2Qgd29yay4gJiM4MjIwOzMmIzgyMjE7IGlzIG1lc3NlbmdlciBSTkEu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBSTkEgaW50cm9ucyBjYW4gYmUgc2VlbiBpbiBudW1iZXLCoCAmIzgyMjA7Mi4mIzgyMjE7IExvb2sgZm9yIFJOQSBpbiB3aGljaCB0aGUgaW50cm9ucyBoYXZlIGJlZW4gc3BsaWNlZCBvdXQu[Qq]
[q] In the diagram below, the protective, 5′ GTP cap is at
[textentry single_char=”true”]
[c]IG c=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwO0cmIzgyMjE7IGlzIHRoZSA1JiM4MjQyOyBHVFAgY2FwLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBHVFAgaXMgYXR0YWNoZWQgdG8gbVJOQSwgYW5kIGl0IGhhcyAzIHBob3NwaGF0ZSBncm91cHMuwqA=[Qq]
[q]In the diagram below, which number represents eukaryotic transcription and translation?
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzImIzgyMjE7IGlzIGV1a2FyeW90aWMgdHJhbnNjcmlwdGlvbiBhbmQgdHJhbnNsYXRpb24u[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBSZW1lbWJlciB0aGF0IGluIGV1a2FyeW90ZXMsIFJOQSBoYXMgdG8gYmUgcHJvY2Vzc2VkIGJlZm9yZSBpdCBjYW4gYmUgdHJhbnNsYXRlZC4gSW4gcHJva2FyeW90ZXMsIHRoYXQmIzgyMTc7cyBub3QgbmVjZXNzYXJ5LsKg[Qq]
[q]In the diagram below, which number represents pre-mRNA?
[textentry single_char=”true”]
[c]IG I=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwO2ImIzgyMjE7IGlzIHByZS1tUk5BLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBQcmUtbVJOQSBjb250YWlucyBpbnRyb25zIGFuZCBsYWNrcyBhIDUmIzgyNDI7IGNhcCBhbmQgYSBwb2x5LUEgdGFpbC7CoA==[Qq]
[q labels = “top”]
[l]introns
[f*] Excellent!
[fx] No. Please try again.
[l]mRNA
[f*] Correct!
[fx] No. Please try again.
[l]poly-A tail
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]pre-mRNA
[f*] Good!
[fx] No. Please try again.
[q labels = “top”]
[l]Alternative splicing
[f*] Great!
[fx] No. Please try again.
[l]pre-mRNA
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]transcription
[f*] Excellent!
[fx] No, that’s not correct. Please try again.
[l]translation
[f*] Great!
[fx] No. Please try again.
[q multiple_choice=”true”] Which of the statements below most closely matches what’s being depicted in the following diagram?
[c]IEV2ZXJ5IEROQSBnZW5lIGhhcyBvbmUgY29ycmVzcG9uZGluZyBwcm90ZWluIHByb2R1Y3Qu[Qq]
[f]IE5vLiBMb29rIGF0ICYjODIyMDtELCYjODIyMTsgJiM4MjIwO0UsJiM4MjIxOyBhbmQgJiM4MjIwO0YsJiM4MjIxOyBhYm92ZS4gVGhyZWUgcHJvdGVpbnMgY29tZSBmcm9tIG9uZSBnZW5lLg==[Qq]
[c]IE11bHRpcGxlIHByb3RlaW5zIGNhbiBiZS Bwcm9kdWNlZCBmcm9tIG9uZSBnZW5lLg==[Qq]
[f]IEV4YWN0bHkuIEJ5IHNodWZmbGluZyBvciBkZWxldGluZyBleG9ucywgYWx0ZXJuYXRpdmUgc3BsaWNpbmcgYWxsb3dzIG11bHRpcGxlIHByb3RlaW5zIHRvIGJlIGNyZWF0ZWQgZnJvbSBvbmUgY29kaW5nIEROQS4=[Qq]
[c]IFByb2thcnlvdGVzIGhhdmUgbW9yZSBmbGV4aWJpbGl0eSBpbiBob3cgZ2VuZXMgYXJlIGV4cHJlc3NlZCB0aGFuIGRvIGV1a2FyeW90ZXM=[Qq]
[f]IE5vLiBUaGF0JiM4MjE3O3MgcHJvYmFibHkgbm90IHRydWUgKGp1c3QgY29uc2lkZXIgdGhlIGFsdGVybmF0aXZlIHNwbGljaW5nIHNob3duIGFib3ZlKSwgYW5kLCBpdCYjODIxNztzIG5vdCBkZXBpY3RlZCBpbiB0aGUgaW1hZ2UgYWJvdmUu[Qq]
[c]TW9kaWZpY2F0aW9uIG9mIEROQSBmcm9tIGNlbGwgdG8gY2VsbCB3aXRoaW4gYSBldWthcnlvdGljIG9yZ2FuaXNtIGNhbiBlbmhhbmNlIHBoZW5vdHlwaWMgZGl2ZXJzaXR5Lg==[Qq]
[f]Tm8uIE5vdGUgdGhhdCBpdCYjODIxNztzIG5vdCB0aGUgRE5BIHRoYXQmIzgyMTc7cyBiZWluZyBtb2RpZmllZC4gVGhlIG1vZGlmaWNhdGlvbiBvY2N1cnMgYWZ0ZXIgdHJhbnNjcmlwdGlvbiwgYW5kIGl0IGludm9sdmVzIFJOQS4=[Qq]
[q] In the diagram below, number 1 is the [hangman].
[c]IHByb21vdGVy[Qq]
[f]IEdvb2Qh[Qq]
[q] In the diagram below, the two deleted RNA segments labeled as f1 and f2 must be [hangman]. Once these are removed (and a few other modifications are made), the result (shown at “3”) is [hangman] RNA.
[c]IGludHJvbnM=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IG1lc3Nlbmdlcg==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] In the diagram below, “d” is the [hangman].
[c]IHByb21vdGVy[Qq]
[f]IENvcnJlY3Qh[Qq]
[x][restart]
[/qwiz]
4.3. Coordinated Control of Gene Expression
[qwiz random = “true” style=”width: 650px !important; min-height: 400px !important;” qrecord_id=”sciencemusicvideosMeister1961-Coordinated Control of Eukaryotic Transcription, APBVP”]
[h]Coordinated Control of Eukaryotic Transcription
[q] In the diagram below, estrogen would be
[textentry single_char=”true”]
[c]IE Y=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtGJiM4MjIxOyByZXByZXNlbnRzIGVzdHJvZ2VuLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIGhvcm1vbmUgdGhhdCYjODIxNztzIGNvbWluZyBmcm9tIG91dHNpZGUgdGhlIGNlbGwsIHRoZW4gZGlmZnVzaW5nIGludG8gdGhlIGN5dG9wbGFzbSB0aHJvdWdoIHRoZSBjZWxsIG1lbWJyYW5lLg==[Qq]
[q] In the diagram below, a mobile estrogen receptor before it has bound to estrogen would be represented by
[textentry single_char=”true”]
[c]IE c=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtHJiM4MjIxOyByZXByZXNlbnRzIGEgbW9iaWxlIGVzdHJvZ2VuIHJlY2VwdG9yIGJlZm9yZSBpdCBiaW5kcyB3aXRoIGVzdHJvZ2VuLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHJlY2VwdG9yIHRoYXQgY2FuIGJpbmQgd2l0aCBlc3Ryb2dlbiAod2hpY2ggaXMgcmVwcmVzZW50ZWQgYnkgJiM4MjIwO0YuJiM4MjIxOyk=[Qq]
[q] In the diagram below, which letter represents the estrogen-receptor complex acting as a transcription factor?
[textentry single_char=”true”]
[c]IG k=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtJJiM4MjIxOyBzaG93cyBhbiBlc3Ryb2dlbi9yZWNlcHRvciBjb21wbGV4IGludGVyYWN0aW5nIHdpdGggRE5BLCBhY3RpbmcgYXMgYSB0cmFuc2NyaXB0aW9uIGZhY3Rvci4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHJlY2VwdG9yIGJvdW5kIHRvIGVzdHJvZ2VuLCBpbnRlcmFjdGluZyB3aXRoIEROQS4=[Qq]
[q] In the diagram below, which letter represents the part that’s translating the transcription product into protein?
[textentry single_char=”true”]
[c]IE w=[Qq]
[f]IENvcnJlY3QhIExldHRlciAmIzgyMjA7TCYjODIyMTsgc2hvd3MgYSByaWJvc29tZS4gdGhlIHJpYm9zb21lIGlzIHRyYW5zbGF0aW5nIFJOQSAodGhlIHRyYW5zY3JpcHRpb24gcHJvZHVjdCkgaW50byBwcm90ZWlu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgdHJhbnNjcmlwdGlvbiBwcm9kdWN0IGlzIFJOQSAoZmlyc3Qgc2hvd24gYXQgJiM4MjIwO0smIzgyMjE7KS4gV2hhdCYjODIxNztzIHRyYW5zbGF0aW5nIHRoZSBSTkEgaW50byBwcm90ZWluPw==[Qq]
[/qwiz]
4.4. Post-transcriptional control of gene expression through micro-RNAs
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Post-transcriptional control of gene expression through micro-RNAs, APBVP”]
[h]Post-transcriptional control of gene expression through micro-RNAs
[q multiple_choice=”true”]
[l]miRNA
[f*] Excellent!
[fx] No. Please try again.
[l]mRNA degradation
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]DNA
[f*] Good!
[fx] No, that’s not correct. Please try again.
[l]precursor miRNA
[f*] Good!
[fx] No. Please try again.
[l]Blocked translation
[f*] Good!
[fx] No. Please try again.
[q multiple_choice=”true”] The type of RNA with a regulatory function is
[c]IG1STkE=[Qq]
[f]IE5vLiBtUk5BIGlzIG1lc3NlbmdlciBSTkEuIEl0IGdldHMgdHJhbnNsYXRlZCBpbnRvIHByb3RlaW4u[Qq]
[c]IG1p Uk5B[Qq]
[f]IENvcnJlY3QuIG1pUk5BIGlzIG1pY3JvUk5BLCBhbmQgaXRzIHJvbGUgaXMgcmVndWxhdGluZyB0cmFuc2xhdGlvbi4=[Qq]
[c]IHRSTkE=[Qq]
[f]IE5vLiB0Uk5BIGlzIHRyYW5zZmVyIFJOQS4gSXRzIHJvbGUgaXMgdG8gYnJpbmcgYW1pbm8gYWNpZHMgdG8gdGhlIHJpYm9zb21lIGR1cmluZyBwcm90ZWluIHN5bnRoZXNpcy4=[Qq]
[q] In the diagram below, a silenced ribosome is represented by the letter
[textentry single_char=”true”]
[c]IE Y=[Qq]
[f]IEdvb2Qgam9iISAmIzgyMjA7RiYjODIyMTsgcmVwcmVzZW50cyBhIHJpYm9zb21lIHRoYXQmIzgyMTc7cyBiZWVuIHNpbGVuY2VkIGJ5IFJOQSBpbnRlcmZlcmVuY2Uu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJbiB0aGlzIGRpYWdyYW0sIG1STkEgaXMgcmVwcmVzZW50ZWQgYXMgYSBibHVlLCBzaW5nbGUgc3RyYW5kLiBXaGF0JiM4MjE3O3MgJiM4MjIwO3JlYWRpbmcmIzgyMjE7IHRoZSBtUk5BPw==[Qq]
[q]The phenomenon shown below is called [hangman] [hangman].
[c]Uk5B[Qq]
[c]aW50ZXJmZXJlbmNl[Qq]
[q]The molecule shown at “E” is called an [hangman] (use an abbreviation, like “tRNA”).
[c]bWlSTkE=[Qq]
[/qwiz]
What’s Next?
Please proceed to our next tutorial: Mutation
