1. All Clades can be taxa…but some (commonly used) taxa are not clades
Any clade (a group derived from a common ancestor) could be a taxon. For example, there’s a name for the clade shown below.
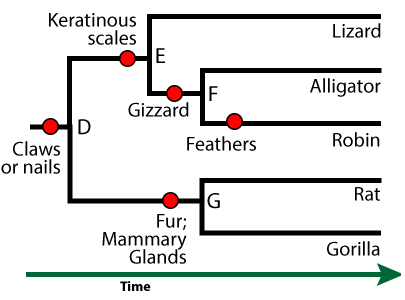
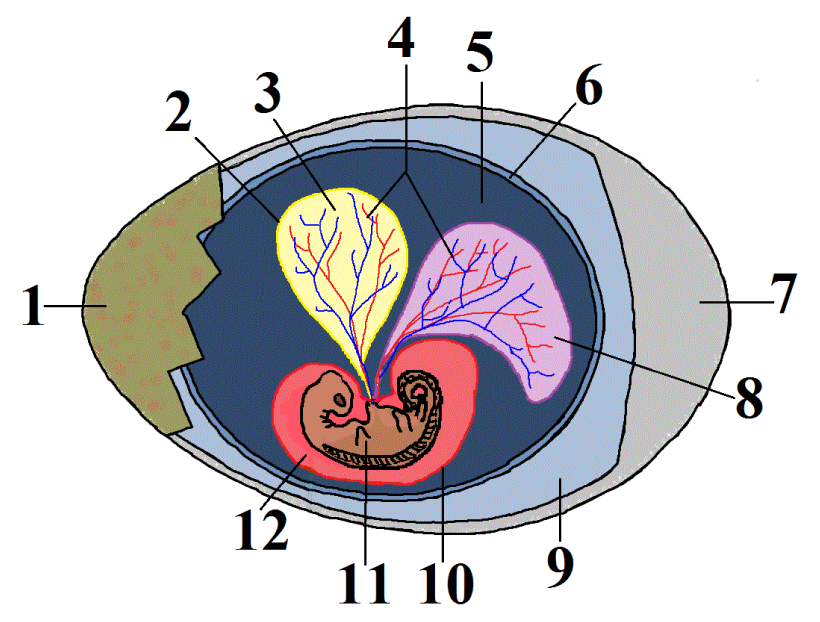
This group is called the amniotes. In addition to possessing the shared derived trait of claws or nails, the females of these species produce a complex egg with an internal, fluid-filled sac that surrounds and nourishes an embryo (shown at 11 at left). This watery internal environment is called an amnion (12). It’s surrounded by an amniotic sac (10).
Note that human beings, like all mammals (including the rat and gorilla in the phylogenetic tree above), are amniotes. Unlike reptiles and birds, mammal eggs, including the amniotic sac and other structures, are kept inside a female’s body.
You’ve probably learned that both alligators and lizards are classified as reptiles. But the taxon we call “reptiles” is not a clade. Remember: a clade includes a common ancestor and all of its descendants.
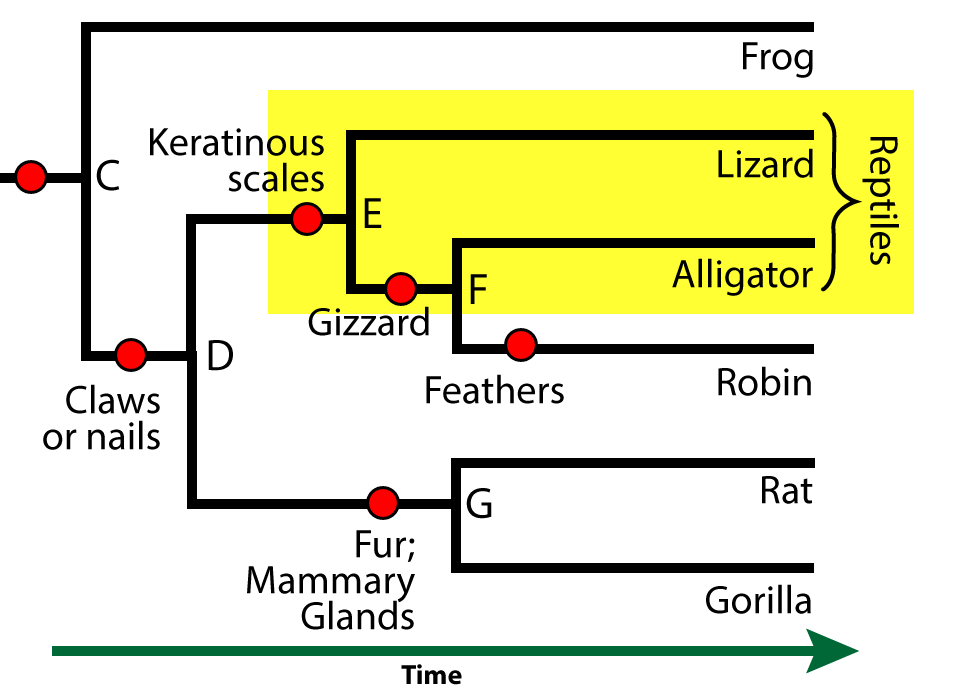 “Reptiles” consists of lizards and alligators (along with other reptiles that aren’t shown here, like snakes), but it excludes birds (with whom the lizards and alligators share a common ancestor).
“Reptiles” consists of lizards and alligators (along with other reptiles that aren’t shown here, like snakes), but it excludes birds (with whom the lizards and alligators share a common ancestor).
Is there a taxon that includes lizards, alligators, and birds? There is one, but it’s not commonly known. It’s called sauropsida, and if you’re interested, you can read more about it on Wikipedia (the link opens in a new browser tab).
Take a look at the groups shown in the cladograms below. Note that all four have the same branching pattern. What’s different are the shaded branches, which indicate different taxa. Some are clades, and some aren’t.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Is it a clade? (v2.0)”]
[h] Is it a clade?
[q labels = “top”]Label the four trees below. If you’re not sure, use trial and error.
[l]Clade
[f*] Good!
[fx] No. Please try again.
[l]Not a clade
[f*] Good!
[fx] No. Please try again.
[q] Taxa 1 and 2 are clades because they include a common ancestor and all of its [hangman]
[c]IGRlc2NlbmRhbnRz[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] Taxon 3 is not a clade because species “c” has a different [hangman] than species “d” and “e.”
[c]IGFuY2VzdG9y[Qq]
[f]IEdvb2Qh[Qq]
[q] The reason why taxon 4 is not a clade is that this taxon is missing one of the [hangman].
[c]IGRlc2NlbmRhbnRz[Qq]
[f]IEdyZWF0IQ==[Qq]
[q multiple_choice=”true”] Based on the discussion above, the taxon “reptiles” would best be represented by which diagram below?
[c]IDEg[Qq][c]IDIg[Qq][c]IDMg[Qq][c]ID Q=
Cg==[Qq][f]IE5vLiAxIGlzIGEgcGVyZmVjdGx5IGdvb2QgY2xhZGUuIFJlcHRpbGVzIGFyZSBub3QgYSBjbGFkZSAoYmVjYXVzZSB0aGV5IGV4Y2x1ZGUgYmlyZHMpLiBXaGljaCBkaWFncmFtIHNob3dzIGFuIGluY29tcGxldGUgY2xhZGU/[Qq]
[f]IE5vLiAyIGlzIGEgcGVyZmVjdGx5IGdvb2QgY2xhZGUuIFJlcHRpbGVzIGFyZSA=bm90IGEgY2xhZGUgKGJlY2F1c2UgdGhleSBleGNsdWRlIGJpcmRzKS4gV2hpY2ggZGlhZ3JhbSBzaG93cyBhbiBpbmNvbXBsZXRlIGNsYWRlPw==[Qq]
[f]IE5vLiBUaGUgcHJvYmxlbSB3aXRoICYjODIyMDszJiM4MjIxOyBpcyB0aGF0IGl0JiM4MjE3O3MgYSB0YXhvbiB3aXRoIG1vcmUgdGhhbiBvbmUgYW5jZXN0b3IuIFJlcHRpbGVzIGFyZSBub3QgYSBjbGFkZSBiZWNhdXNlIHRoZXkgZXhjbHVkZSBvbmUgb2YgdGhlIGRlc2NlbmRhbnRzIChiaXJkcykuIFdoaWNoIGRpYWdyYW0gc2hvd3MgYW4gaW5jb21wbGV0ZSBjbGFkZT8=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzQmIzgyMjE7IGlzIG5vdCBhIGNsYWRlIGluIHRoZSBzYW1lIHdheSB0aGF0IHJlcHRpbGVzIGFyZSBub3QgYSBjbGFkZS4gSXQmIzgyMTc7cyBhIGdyb3VwIHdpdGggYSBjb21tb24gYW5jZXN0b3IsIGJ1dCBub3QgYWxsIG9mIHRoZSBkZXNjZW5kYW50cy4gSW4gdGhlIHNhbWUgd2F5LCB0aGUgcmVwdGlsZSB0YXhvbiBsZWF2ZXMgb3V0IGJpcmRzLg==[Qq]
[/qwiz]
2. In phylogenetic analysis, watch out for analogous features
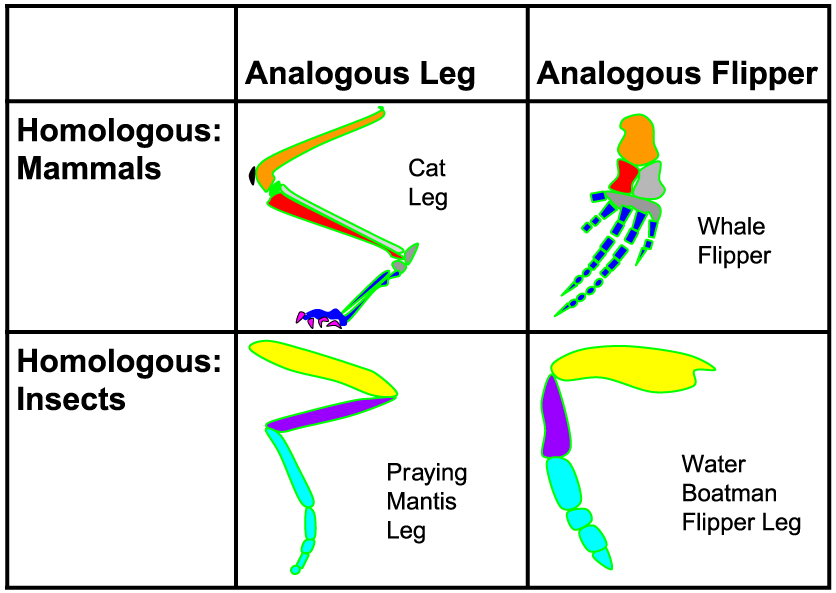
In terms of interpreting and creating phylogenies, not all inherited characters are of equal value. Characters are only useful if they’re homologous (derived from a common ancestor). But if they’re analogous traits — ones that come about from convergent evolution — they can put you off track in determining a correct phylogeny.
Use the diagram on the right to review the difference between homology and analogy.
- HOMOLOGY: The leg of a cat and the flipper of a whale are homologous: the underlying structure derives from a common ancestor. The same is true for the leg of a praying mantis and a water boatman.
- ANALOGY: The leg of the cat and the leg of the mantis are analogous features that emerged through convergent evolution. The same is true of the whale’s flipper and the water boatman’s leg.
[qwiz style=”width: 600px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-Analogy, Homology, and Phylogeny (v2.0)”]
[h] Analogy, Homology, and Phylogeny
[i] Let’s say that you were assigned the task of figuring out a phylogenetic tree for the following five organisms: a rat, a bat, a bird, a frog, and a fish.
[q labels = “top”]In the following character table, use “X” to indicate a trait’s presence, and a “0” to indicate its absence. Note that fish and frogs have simple eggs (not amniotic eggs).
| Shared Characters | Rat | Bat | Bird | Frog | Fish |
| Vertebral column | ___ | ___ | ___ | ___ | ___ |
| Four limbs | ___ | ___ | ___ | ___ | ___ |
| Wings | ___ | ___ | ___ | ___ | ___ |
| Amniotic egg | ___ | ___ | ___ | ___ | ___ |
[l]X
[f*] Correct!
[fx] No, that’s not correct. Please try again.
[l]0
[f*] Great!
[fx] No, that’s not correct. Please try again.
[q labels = “top”]Now, use the character table you just created to complete the phylogenetic tree below. Note: this phylogeny is wrong: we’re making it to prove a point.
[l]Amniotic sac
[f*] Excellent!
[fx] No, that’s not correct. Please try again.
[l]Bird
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]Fish
[f*] Excellent!
[fx] No, that’s not correct. Please try again.
[l]Four limbs
[f*] Good!
[fx] No, that’s not correct. Please try again.
[l]Frog
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]Rat
[f*] Correct!
[fx] No. Please try again.
[l]Wings
[f*] Excellent!
[fx] No. Please try again.
[q] The problem with the phylogenetic tree below is that one of the characters, [hangman], is NOT a shared [hangman] feature. Rather, it’s an [hangman] trait that resulted from convergent evolution (adaptation to a flying niche).
[c]IHdpbmdz[Qq]
[f]IEdyZWF0IQ==[Qq]
[c]IGRlcml2ZWQ=[Qq]
[f]IEdvb2Qh[Qq]
[c]IGFuYWxvZ291cw==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q]Let’s try to analyze these organisms using improved characters. As opposed to wings (an analogous trait) in birds and bats, let’s use hair and feathers instead.
| Shared Characters | Rat | Bat | Bird | Frog | Fish |
| Vertebral column | ___ | ___ | ___ | ___ | ___ |
| Four limbs | ___ | ___ | ___ | ___ | ___ |
| Hair | ___ | ___ | ___ | ___ | ___ |
| Amniotic egg | ___ | ___ | ___ | ___ | ___ |
| Feathers | ___ | ___ | ___ | ___ | ___ |
[l]X
[f*] Good!
[fx] No, that’s not correct. Please try again.
[l]0
[f*] Excellent!
[fx] No. Please try again.
[q labels = “top”]Based on the character table below, draw a new phylogenetic tree.
[l]Amniotic sac
[f*] Great!
[fx] No. Please try again.
[l]Bat
[f*] Excellent!
[fx] No, that’s not correct. Please try again.
[l]Bird
[f*] Good!
[fx] No, that’s not correct. Please try again.
[l]Feathers
[f*] Correct!
[fx] No. Please try again.
[l]Fish
[f*] Excellent!
[fx] No. Please try again.
[l]Four limbs
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]Frog
[f*] Excellent!
[fx] No. Please try again.
[l]Hair
[f*] Excellent!
[fx] No, that’s not correct. Please try again.
[q] The advantage of tree 1 below is that it only uses traits that are shared [hangman] characters. The takeaway: when constructing phylogenetic trees, make sure you use traits that are [hangman] (from common ancestry), as opposed to [hangman] (from convergent evolution).
[c]IGRlcml2ZWQ=[Qq]
[f]IEdvb2Qh[Qq]
[c]IGhvbW9sb2dvdXM=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IGFuYWxvZ291cw==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] The phylogenetic tree below shows a variety of mammals, along with their ability to synthesize vitamin C. Lineages shown in black can synthesize vitamin C. Those in gray can’t, and must obtain vitamin C from their environment. Based on the pattern in the diagram, the inability of primates (owl marmosets to humans) is a [hangman] [hangman] feature. But, the fact that guinea pigs can’t synthesize vitamin C is the result of [hangman] evolution. It’s an [hangman] trait.
[c]IHNoYXJlZA==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IGRlcml2ZWQ=[Qq]
[f]IEdyZWF0IQ==[Qq]
[c]IGNvbnZlcmdlbnQ=[Qq]
[f]IENvcnJlY3Qh[Qq]
[c]IGFuYWxvZ291cw==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[/qwiz]
3. Phylogeny and “Molecular Clocks”
In all cladograms and phylogenetic trees, there’s an underlying assumption of the passage of time.
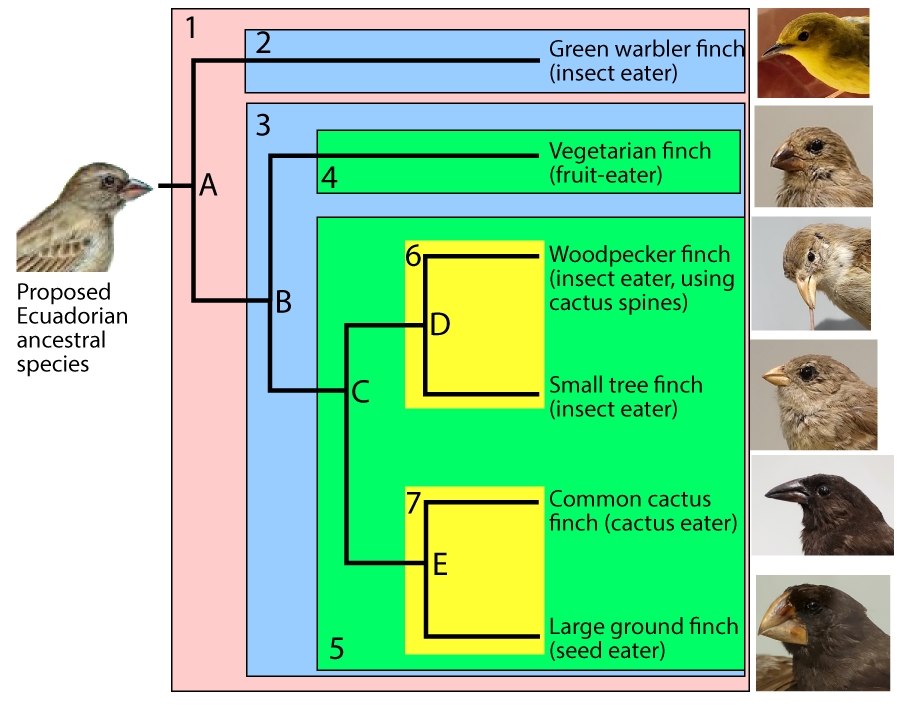
For example, in the phylogenetic tree above, you can assume that clade E emerged more recently than clade C, which emerged more recently than clade B. But there’s no explicit scale.
In other phylogenetic trees, the branch lengths are proportional to the passage of time.
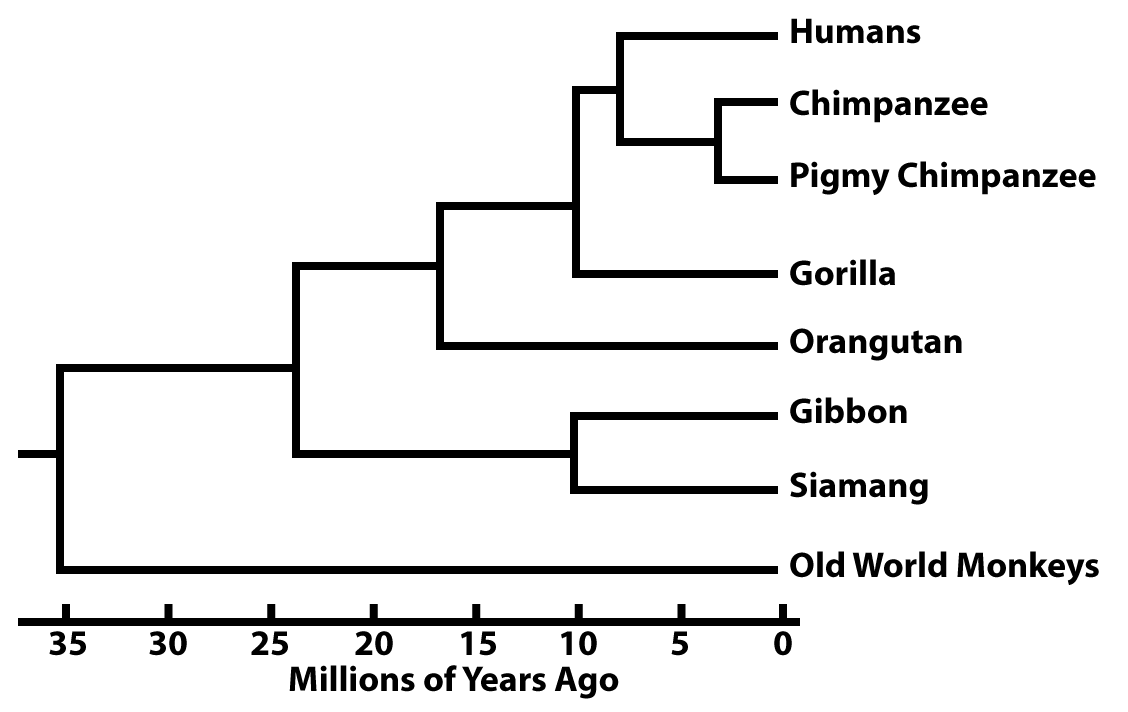
These branch points can be dated from fossil evidence. But increasingly they’re dated based on what are called molecular clocks.
The basic idea behind molecular clocks is that for any specific protein or nucleic acid, the rate of change caused by accumulated mutations is constant over time. If you can calibrate the amount of change in a gene or protein to when two species split apart (a node in the tree), then you can
- Determine a rate of change in that gene or protein,
- Use that rate to determine when other species split in the tree split apart.
Here’s an imaginary example. Imagine a specific sequence of DNA that’s 100 nucleotides long, and all of the primates shown above share it. In Gibbons and Siamangs, that stretch of DNA is identical except for 10 mutations. Based on fossil evidence, we learn that the split between Gibbons and Siamangs occurred 10 million years ago.
That means that the rate of change in that nucleotide sequence is
- 10 mutations/10 million years, or
- one mutation/million years.
If we find that there are six nucleotide differences in the same stretch of DNA that’s shared by humans and chimps, then a little algebra tells us that humans and chimps would have had a common ancestor about 6 million years ago.
[qwiz style=”width: 600px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-Molecular Clocks (v2.0)”]
[h] Molecular Clocks
[i]A widely used and highly conserved protein that’s used to determine divergence dates is hemoglobin, the oxygen-carrying protein found in the red blood cells of almost all vertebrates. Hemoglobin consists of two pairs of protein chains. Two of these chains consist of 141 amino acids, and two have 146 amino acids.
The rate of mutation in hemoglobin has been calibrated with changes in the fossil record, allowing scientists to produce the graph below.
[q multiple_choice=”true”] There are 62 amino acid differences between human and frog hemoglobin. Use the graph below to determine when humans and frogs most recently shared a common ancestor.
[c]IDEwMCBtaWxsaW9uIHllYXJz[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgaG93IHRvIHByb2NlZWQuIElkZW50aWZ5IDYyIG9uIHRoZSBZLWF4aXMuIE5vdyBmaW5kIHdoZXJlIDYyIGludGVyY2VwdHMgdGhlIG1vbGVjdWxhciBjbG9jayBsaW5lLCBhbmQgdGhlbiBmaW5kIHRoZSBjb3JyZXNwb25kaW5nIHRpbWUgb24gdGhlIFgtYXhpcy4=[Qq]
[c]IDI1MCBtaWxsaW9uIHllYXJz[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgaG93IHRvIHByb2NlZWQuIElkZW50aWZ5IDYyIG9uIHRoZSBZLWF4aXMuIE5vdyBmaW5kIHdoZXJlIDYyIGludGVyY2VwdHMgdGhlIG1vbGVjdWxhciBjbG9jayBsaW5lLCBhbmQgdGhlbiBmaW5kIHRoZSBjb3JyZXNwb25kaW5nIHRpbWUgb24gdGhlIFgtYXhpcy4=[Qq]
[c]IDM1MCBtaWxs aW9uIHllYXJz[Qq]
[f]RXhjZWxsZW50LiBVc2luZyB0aGUgaGVtb2dsb2JpbiBtb2xlY3VsYXIgY2xvY2ssIHlvdSBjYW4gZXN0aW1hdGUgdGhhdCB0aGUgbGFzdCBjb21tb24gYW5jZXN0b3Igb2YgaHVtYW5zIGFuZCBmcm9ncyBleGlzdGVkIGFib3V0IDM1MCBtaWxsaW9uIHllYXJzIGFnby4=[Qq]
[c]IDUwMCBtaWxsaW9uIHllYXJz[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgaG93IHRvIHByb2NlZWQuIElkZW50aWZ5IDYyIG9uIHRoZSBZIGF4aXMuIE5vdyBmaW5kIHdoZXJlIDYyIGludGVyY2VwdHMgdGhlIG1vbGVjdWxhciBjbG9jayBsaW5lLCBhbmQgdGhlbiBmaW5kIHRoZSBjb3JyZXNwb25kaW5nIHRpbWUgb24gdGhlIFgtYXhpcy4=[Qq]
[q multiple_choice=”true”] Cytochrome c is also used as a molecular clock.
Humans and fruit flies have 29 differences in the amino acids making up their cytochrome c. Based on the graph below, which of the following is closest to the time when humans and fruit flies last shared a common ancestor?
[c]IDEwMCBtaWxsaW9uIHllYXJz[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgaG93IHRvIHByb2NlZWQuIElkZW50aWZ5IDI5IG9uIHRoZSBZIGF4aXMuIE5vdyBmaW5kIHdoZXJlIDI5IGludGVyY2VwdHMgdGhlIG1vbGVjdWxhciBjbG9jayBsaW5lLCBhbmQgdGhlbiBmaW5kIHRoZSBjb3JyZXNwb25kaW5nIHRpbWUgb24gdGhlIFgtYXhpcy4=[Qq]
[c]IDI1MCBtaWxsaW9uIHllYXJz[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgaG93IHRvIHByb2NlZWQuIElkZW50aWZ5IDI5IG9uIHRoZSBZIGF4aXMuIE5vdyBmaW5kIHdoZXJlIDI5IGludGVyY2VwdHMgdGhlIG1vbGVjdWxhciBjbG9jayBsaW5lLCBhbmQgdGhlbiBmaW5kIHRoZSBjb3JyZXNwb25kaW5nIHRpbWUgb24gdGhlIFgtYXhpcy4=[Qq]
[c]IDQwMCBtaWxsaW9uIHllYXJz[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgaG93IHRvIHByb2NlZWQuIElkZW50aWZ5IDI5IG9uIHRoZSBZIGF4aXMuIE5vdyBmaW5kIHdoZXJlIDI5IGludGVyY2VwdHMgdGhlIG1vbGVjdWxhciBjbG9jayBsaW5lLCBhbmQgdGhlbiBmaW5kIHRoZSBjb3JyZXNwb25kaW5nIHRpbWUgb24gdGhlIFgtYXhpcy4=[Qq]
[c]IDUwMCBtaWxs aW9uIHllYXJz[Qq]
[f]IEV4Y2VsbGVudC4gSWYgdGhlcmUgYXJlIDI5IGFtaW5vIGFjaWQgZGlmZmVyZW5jZXMgYmV0d2VlbiBodW1hbiBhbmQgZnJ1aXQgZmx5IGN5dG9jaHJvbWUgYywgdGhlbiB0aGVpciBkaXZlcmdlbmNlIGRhdGUgc2hvdWxkIGJlIGFib3V0IDUwMCBtaWxsaW9uIHllYXJzIGFnbw==[Qq]
[/qwiz]
3. Phylogenetic Trees, Checking Understanding
This module on classification and phylogenetic trees has covered
- Why we need to classify living things.
- How we name species using binomial nomenclature.
- The classification categories used to classify species into taxa above the species level.
- The three domains
- How a key goal of taxonomy is to create taxa that are clades.
- How to analyze lineages, nodes, and common ancestors within phylogenetic trees.
- How to use phylogenetic trees and cladograms to determine how closely related taxa are.
- How to use shared derived features to create phylogenetic trees and cladograms.
- The difference between shared derived features and ancestral features.
- The types of evidence used to construct phylogenetic trees: anatomical/morphological or molecular (proteins and nucleic acid sequences).
- How data from DNA and proteins can be used to construct molecular clocks.
Use the quiz below to check how well you’ve mastered this material.
[qwiz style=”width: 700px !important; min-height: 450px !important;” qrecord_id=”sciencemusicvideosMeister1961-Phylogenetic Trees: Checking Understanding”]
[h] Phylogenetic Trees: Checking Understanding
[i]
[q multiple_choice=”true”] In the phylogenetic tree below, the shared derived trait of clade “F” is (a)
[c]IEtlcmF0aW5vdXMgc2NhbGVz[Qq]
[f]IE5vLiBGb3IgY2xhZGUgJiM4MjIwO0YsJiM4MjIxOyBrZXJhdGlub3VzIHNjYWxlcyB3b3VsZCBiZSBhbiBhbmNlc3RyYWwgZmVhdHVyZS4gQSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIGlzIG9uZSB0aGF0IHNldHMgdGhlIG1lbWJlcnMgb2YgYSBjbGFkZSBhcGFydCBmcm9tIG90aGVyIGNsYWRlcy4=[Qq]
[c]IEdpen phcmQ=[Qq]
[f]IEV4Y2VsbGVudC4gQSBnaXp6YXJkIHNldHMgdGhlIG1lbWJlcnMgb2YgYSBjbGFkZSAmIzgyMjA7RiYjODIyMTsgYXBhcnQgZnJvbSBvdGhlciBjbGFkZXMsIG1ha2luZyBpdCB0aGVpciBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlLg==[Qq]
[c]IEZlYXRoZXJz[Qq]
[f]IE5vLiBPbmx5IHNvbWUgbWVtYmVycyBvZiBjbGFkZSAmIzgyMjA7ZiYjODIyMTsgaGF2ZSBmZWF0aGVycy4gQSBzaGFyZWQgZGVyaXZlZCBmZWF0dXJlIGlzIG9uZSB0aGF0IGlzIHNoYXJlZCBieSBhbGwgbWVtYmVycyBvZiBhIGNsYWRlLCBhbmQgd2hpY2ggc2V0cyB0aGUgbWVtYmVycyBvZiBhIGNsYWRlIGFwYXJ0IGZyb20gb3RoZXIgY2xhZGVzLiBGaW5kIGEgZmVhdHVyZSB0aGF0IGZpdHMgdGhvc2UgcmVxdWlyZW1lbnRzLg==[Qq]
[q multiple_choice=”true”] In the phylogenetic tree below, an ancestral trait for clade “F” would be
[c]IEtlcmF0aW5v dXMgc2NhbGVz[Qq]
[f]IEV4Y2VsbGVudC4gRm9yIGNsYWRlICYjODIyMDtGLCYjODIyMTsgJiM4MjIwO2tlcmF0aW5vdXMgc2NhbGVzJiM4MjIxOyBpcyBhbiBhbmNlc3RyYWwgZmVhdHVyZS4gSXQgd2FzIGluaGVyaXRlZCBmcm9tIGEgY29tbW9uIGFuY2VzdG9yLCBidXQgbm90IGZyb20gYSBjbG9zZSBlbm91Z2ggYW5jZXN0b3IgdG8gZGlzdGluZ3Vpc2ggdGhpcyBjbGFkZSBmcm9tIG90aGVyIGNsYWRlcy4=[Qq]
[c]IEdpenphcmQ=[Qq]
[f]IE5vLiBBIGdpenphcmQgc2V0cyB0aGUgbWVtYmVycyBvZiBhIGNsYWRlICYjODIyMDtGJiM4MjIxOyBhcGFydCBmcm9tIG90aGVyIGNsYWRlcywgbWFraW5nIGl0IGEgc2hhcmVkIGRlcml2ZWQgZmVhdHVyZS4gQW4gYW5jZXN0cmFsIHRyYWl0IGlzIGFsc28gaW5oZXJpdGVkIGZyb20gYSBjb21tb24gYW5jZXN0b3IsIGJ1dCBpdCYjODIxNztzIG5vdCBhIHRyYWl0IHRoYXQgZGlzdGluZ3Vpc2hlcyB0aGlzIGNsYWRlIGZyb20gb3RoZXIgY2xhZGVzLg==[Qq]
[c]IEZlYXRoZXJz[Qq]
[f]IE5vLiBPbmx5IHNvbWUgbWVtYmVycyBvZiBjbGFkZSAmIzgyMjA7ZiYjODIyMTsgaGF2ZSBmZWF0aGVycywgbWFraW5nIHRoYXQgZmVhdHVyZSBuZWl0aGVyIGEgc2hhcmVkIGRlcml2ZWQgdHJhaXQgbm9yIGFuIGFuY2VzdHJhbCB0cmFpdC4=
Cg==[Qq]
[q] Which of the trees below depicts a taxon that has more than one ancestor?
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IE5pY2Ugam9iISAmIzgyMjA7MyYjODIyMTtoYXMgbW9yZSB0aGFuIG9uZSBhbmNlc3Rvci4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJbiB0YXhhIDEgYW5kIDIsIHRoZXJlJiM4MjE3O3MgYSBzaW5nbGUgbm9kZSB0aGF0IGxlYWRzIHRvIGFsbCBvZiB0aGUgZGVzY2VuZGFudHMgd2l0aGluIHRoYXQgY2xhZGUuIFdoaWNoIGRpYWdyYW0gc2hvd3MgbW9yZSB0aGFuIG9uZSBub2RlIGJlbmVhdGggdGhlIGRlc2NlbmRhbnRzPw==[Qq]
[q] Which of the trees below depicts a taxon that’s an incomplete clade?
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IEdvb2Qgam9iISAmIzgyMjA7NCYjODIyMTsgaXMgYW4gaW5jb21wbGV0ZSBjbGFkZSAoYmVjYXVzZSBpdCYjODIxNztzIG1pc3Npbmcgb25lIG9mIGl0cyBkZXNjZW5kYW50cyku[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBBIGxvb2sgZm9yIGEgdGF4b24gaW4gd2hpY2ggdGhlIG1lbWJlcnMgc2hhcmUgdGhlIHNhbWUgY29tbW9uIGFuY2VzdG9yLCBidXQgbm90IGFsbCBvZiB0aGUgZGVzY2VuZGFudHMgYXJlIHBhcnQgb2YgdGhlIGNsYWRlLg==[Qq]
[q] If a taxon consists of an ancestral species and all of its descendants, then it can be described in one word: a [hangman].
[c]IGNsYWRl[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] The problem with a taxon that included only alligators and lizards is that this taxon would not be a [hangman]. That’s because its common ancestor at letter E was also the ancestor of birds
[c]IGNsYWRl[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] A key principle of evolutionary classification is the more closely related two taxa are, the more recently they shared a [hangman] [hangman].
[c]IGNvbW1vbg==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IGFuY2VzdG9y[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] In the diagram below, the most recent common ancestor of A and B would be
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IEdyZWF0ISAmIzgyMjA7MSYjODIyMTsgaXMgdGhlIG1vc3QgcmVjZW50IGNvbW1vbiBhbmNlc3RvciBvZiBBIGFuZCBCLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIHRoZSBub2RlIHRoYXQgbW9zdCBpbW1lZGlhdGVseSBjb25uZWN0cyAmIzgyMjA7QSYjODIyMTsgYW5kICYjODIyMDtCLiYjODIyMTsgVGhhdCYjODIxNztzIHRoZWlyIG1vc3QgcmVjZW50IGNvbW1vbiBhbmNlc3Rvci7CoA==[Qq]
[q] In the diagram below, the most recent common ancestor of A, B, C, D, and E would be
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEF3ZXNvbWUhICYjODIyMDsyJiM4MjIxOyBpcyB0aGUgbW9zdCByZWNlbnQgY29tbW9uIGFuY2VzdG9yIG9mIEEsIEIsIEMsIEQsIGFuZCBFLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBNb3ZlIGJhY2sgaW4gdGltZSBmcm9tIEEsIEIsIEMsIEQsIGFuZCBFIGFuZCBmaW5kIGEgcG9pbnQgd2hlcmUgYWxsIHRoZWlyIGxpbmVhZ2VzIGNvbnZlcmdlLiBUaGF0JiM4MjE3O3MgdGhlaXIgY29tbW9uIGFuY2VzdG9yLsKg[Qq]
[q] If you made up a taxon that included A, B, and C, that taxon would not be a [hangman].
[c]IGNsYWRl[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q multiple_choice=”true”] According to the diagram below, which of the following statements is true?
[c]IEh1bWFucyBldm9sdmVkIGZyb20gT2xkIFdvcmxkIG1vbmtleXM=[Qq]
[f]IE5vLiAmIzgyMjA7RXZvbHZlZCBmcm9tJiM4MjIxOyBtZWFucywgbW9yZSBvciBsZXNzLCAmIzgyMjA7YXJlIGRlc2NlbmRlZCBmcm9tLiYjODIyMTsgSHVtYW5zIGFuZCBPbGQgV29ybGQgTW9ua2V5cyBhcmUgZXZvbHV0aW9uYXJ5IA==Y291c2lucw==LiBZb3VyIGNvdXNpbnMgYXJlbiYjODIxNzt0IHlvdXIgYW5jZXN0b3JzLg==[Qq]
[c]IEh1bWFucyBhbmQgbmV3IHdvcmxkIG1vbmtl eXMgYmVsb25nIHRvIHRoZSBzYW1lIGNsYWRl[Qq]
[f]IFRoYXQmIzgyMTc7cyBjb3JyZWN0LiBIdW1hbnMgYW5kIG5ldyB3b3JsZCBtb25rZXlzIHNoYXJlIGEgY29tbW9uIGFuY2VzdG9yIGFuZCBhcmUgdGhlcmVmb3JlIG1lbWJlcnMgb2YgdGhlIHNhbWUgY2xhZGUu[Qq]
[q] The study of evolutionary history and relationships is known as [hangman]. (hint: it begins with a “p”).
[c]IHBoeWxvZ2VueQ==[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] The two-word name for every species, such as Homo sapiens or Bos taurus, is known as a [hangman].
[c]IGJpbm9taWFs[Qq]
[f]IEdvb2Qh[Qq]
[q] In a species name like Turdus migratorius (the American robin), Turdus denotes the [hangman], while migratorius indicates the [hangman].
[c]IGdlbnVz[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IHNwZWNpZXM=[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] Of the classification categories Kingdom, class, and family, the most general one is [hangman], while the most exclusive/specific one is [hangman]
[c]IGtpbmdkb20=[Qq]
[c]ZmFtaWx5[Qq]
[q multiple_choice=”true”] Any named group of organisms is a
[c]IHRh eG9u[Qq]
[f]IENvcnJlY3Qh[Qq]
[c]IHNwZWNpZXM=[Qq]
[f]IE5vLiBXaGlsZSBhIHNwZWNpZXMgaXMgYSB0YXhvbiwgbm90IGFsbCB0YXhhIGFyZSBzcGVjaWVzLsKg[Qq]
[c]IGNsYWRl[Qq]
[f]IE5vLiBNYW55IG5hbWVkIGdyb3VwcyBvZiBvcmdhbmlzbXMgKGxpa2UgJiM4MjIwO3dhdGVyZm93bCwmIzgyMjE7ICYjODIyMDttYXJpbmUgbWFtbWFscywmIzgyMjE7IG9yICYjODIyMDtyZXB0aWxlcyYjODIyMTspIGFyZSBub3QgY2xhZGVzLsKg[Qq]
[q] Letters “A,” “B,” and “C” are points where [hangman] events occurred. They’re called [hangman].
[c]IHNwZWNpYXRpb24=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IG5vZGVz[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] The basic idea behind molecular clocks is that for any specific protein or nucleic acid, the rate of change caused by accumulated [hangman] is constant over time.
[c]IG11dGF0aW9ucw==[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] Because mutations accumulate in proteins or nucleic acids at a relatively constant rate, you can infer that mice share a more recent common ancestor with [hangman] and gorillas than they do with chickens.
[c]IG1vbmtleXM=[Qq]
[f]IEdvb2Qh[Qq]
[q] In the diagram below, the shared derived trait of clade G is indicated by number
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzQmIzgyMjE7IHNob3dzIHRoZSBzaGFyZWQgZGVyaXZlZCB0cmFpdCBvZiByYXRzIGFuZCBnb3JpbGxhcyAobWVtYmVycyBvZiB0aGUgbWFtbWFsIGNsYWRlKQ==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHRyYWl0IHRoYXQmIzgyMTc7cyB1bmlxdWUgZm9yIHRoZSBjbGFkZSBpbiB3aGljaCBib3RoIHJhdHMgYW5kIGdvcmlsbGFzIGFyZSBmb3VuZC4=[Qq]
[q] In the diagram below, the common ancestor of lizards, alligators, robins, rats, and gorillas is indicated by letter
[textentry single_char=”true”]
[c]IE Q=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwO0QmIzgyMjE7IGlzIHRoZSBjb21tb24gYW5jZXN0b3Igb2YgdGhlIGNsYWRlIHRoYXQgdW5pdGVzIGxpemFyZHMsIGFsbGlnYXRvcnMsIHJvYmlucywgcmF0cywgYW5kIGdvcmlsbGFzLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvbGxvdyB0aGUgbGluZWFnZXMgbGVhZGluZyB0byBsaXphcmRzLCBhbGxpZ2F0b3JzLCByb2JpbnMsIHJhdHMsIGFuZCBnb3JpbGxhcyBiYWNrIGluIHRpbWUgdW50aWwgdGhleSBtZWV0IGluIGEgY29tbW9uIGFuY2VzdG9yLg==[Qq]
[q] In the diagram below, what number indicates the most recent ancestral trait of lizards, alligators, robins, rats, and gorillas?
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzImIzgyMjE7IGlzIHRoZSBtb3N0IHJlY2VudCBhbmNlc3RyYWwgdHJhaXQgb2YgdGhlIGNsYWRlIHRoYXQgdW5pdGVzIGxpemFyZHMsIGFsbGlnYXRvcnMsIHJvYmlucywgcmF0cywgYW5kIGdvcmlsbGFzICh0aGUgc2hhcmVkIGRlcml2ZWQgdHJhaXQgaXMgJiM4MjIwOzMuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIHRoZSBzaGFyZWQgZGVyaXZlZCB0cmFpdCBvciB0aGUgY2xhZGUgdGhhdCB1bml0ZXMgbGl6YXJkcywgYWxsaWdhdG9ycywgcm9iaW5zLCByYXRzLCBhbmQgZ29yaWxsYXMuIFRoZW4gZmluZCB0aGUgdHJhaXQgdGhhdCB3b3VsZCBpbmNsdWRlIHRoZSBvdXRncm91cCBvZiB0aGF0IGNsYWRlICh3aGljaCBpcyB0aGUgZnJvZyku[Qq]
[/qwiz]
What now?
This tutorial ends AP Bio Topic 7.9, Phylogeny and Classification. To continue with AP Bio Unit 7, use this link to go to the AP Bio Unit 7 Main Menu.
